Medicinal chemistry advances in targeting class I histone deacetylases
- PMID: 37711592
- PMCID: PMC10497394
- DOI: 10.37349/etat.2023.00166
Medicinal chemistry advances in targeting class I histone deacetylases
Abstract
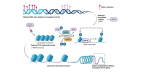
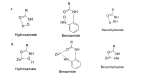
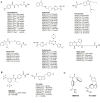
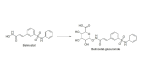
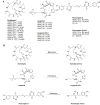
Histone deacetylases (HDACs) are a class of zinc (Zn)-dependent metalloenzymes that are responsible for epigenetic modifications. HDACs are largely associated with histone proteins that regulate gene expression at the DNA level. This tight regulation is controlled by acetylation [via histone acetyl transferases (HATs)] and deacetylation (via HDACs) of histone and non-histone proteins that alter the coiling state of DNA, thus impacting gene expression as a downstream effect. For the last two decades, HDACs have been studied extensively and indicated in a range of diseases where HDAC dysregulation has been strongly correlated with disease emergence and progression-most prominently, cancer, neurodegenerative diseases, HIV, and inflammatory diseases. The involvement of HDACs as regulators in these biochemical pathways established them as an attractive therapeutic target. This review summarizes the drug development efforts exerted to create HDAC inhibitors (HDACis), specifically class I HDACs, with a focus on the medicinal chemistry, structural design, and pharmacology aspects of these inhibitors.
Keywords: Histone deacetylases; cap group; epigenetic regulation; medicinal chemistry; small-molecule inhibitors; zinc-binding group.
© The Author(s) 2023.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



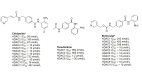


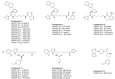
Similar articles
-
Recent advances in class IIa histone deacetylases research.Bioorg Med Chem. 2019 Nov 15;27(22):115087. doi: 10.1016/j.bmc.2019.115087. Epub 2019 Sep 6. Bioorg Med Chem. 2019. PMID: 31561937 Review.
-
Histone Deacetylase Inhibitors, Intrinsic and Extrinsic Apoptotic Pathways, and Epigenetic Alterations of Histone Deacetylases (HDACs) in Hepatocellular Carcinoma.Iran J Pharm Res. 2021 Summer;20(3):324-336. doi: 10.22037/ijpr.2021.115105.15197. Iran J Pharm Res. 2021. PMID: 34903992 Free PMC article.
-
The emerging roles of HDACs and their therapeutic implications in cancer.Eur J Pharmacol. 2022 Sep 15;931:175216. doi: 10.1016/j.ejphar.2022.175216. Epub 2022 Aug 18. Eur J Pharmacol. 2022. PMID: 35988787 Review.
-
Inhibition of class II histone deacetylases in the spinal cord attenuates inflammatory hyperalgesia.Mol Pain. 2010 Sep 7;6:51. doi: 10.1186/1744-8069-6-51. Mol Pain. 2010. PMID: 20822541 Free PMC article.
-
Characterization of the Expression and Role of Histone Acetylation and Deacetylation in Dental Pulp Cells.Methods Mol Biol. 2023;2588:279-293. doi: 10.1007/978-1-0716-2780-8_17. Methods Mol Biol. 2023. PMID: 36418694
Cited by
-
Histone deacetylase inhibitors for leukemia treatment: current status and future directions.Eur J Med Res. 2024 Oct 26;29(1):514. doi: 10.1186/s40001-024-02108-8. Eur J Med Res. 2024. PMID: 39456044 Free PMC article. Review.
-
SIN3B Loss Heats up Cold Tumor Microenvironment to Boost Immunotherapy in Pancreatic Cancer.Adv Sci (Weinh). 2024 Nov;11(43):e2402244. doi: 10.1002/advs.202402244. Epub 2024 Sep 24. Adv Sci (Weinh). 2024. PMID: 39316363 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
