Contact sites between endoplasmic reticulum sheets and mitochondria regulate mitochondrial DNA replication and segregation
- PMID: 37534187
- PMCID: PMC10391914
- DOI: 10.1016/j.isci.2023.107180
Contact sites between endoplasmic reticulum sheets and mitochondria regulate mitochondrial DNA replication and segregation
Abstract
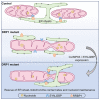
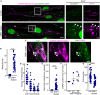
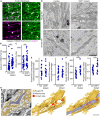
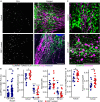
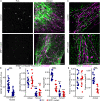
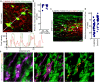
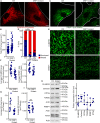
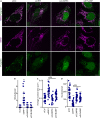
Mitochondria are multifaceted organelles crucial for cellular homeostasis that contain their own genome. Mitochondrial DNA (mtDNA) replication is a spatially regulated process essential for the maintenance of mitochondrial function, its defect causing mitochondrial diseases. mtDNA replication occurs at endoplasmic reticulum (ER)-mitochondria contact sites and is affected by mitochondrial dynamics: The absence of mitochondrial fusion is associated with mtDNA depletion whereas loss of mitochondrial fission causes the aggregation of mtDNA within abnormal structures termed mitobulbs. Here, we show that contact sites between mitochondria and ER sheets, the ER structure associated with protein synthesis, regulate mtDNA replication and distribution within mitochondrial networks. DRP1 loss or mutation leads to modified ER sheets and alters the interaction between ER sheets and mitochondria, disrupting RRBP1-SYNJ2BP interaction. Importantly, mtDNA distribution and replication were rescued by promoting ER sheets-mitochondria contact sites. Our work identifies the role of ER sheet-mitochondria contact sites in regulating mtDNA replication and distribution.
Keywords: Biochemistry; Biological sciences; Cell biology.
© 2023 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
ER-mitochondria contact sites in mitochondrial DNA dynamics, maintenance, and distribution.Int J Biochem Cell Biol. 2024 Jan;166:106492. doi: 10.1016/j.biocel.2023.106492. Epub 2023 Nov 4. Int J Biochem Cell Biol. 2024. PMID: 37931682 Review.
-
ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells.Science. 2016 Jul 15;353(6296):aaf5549. doi: 10.1126/science.aaf5549. Science. 2016. PMID: 27418514 Free PMC article.
-
Interaction Between Mitochondrial DNA Variants and Mitochondria/Endoplasmic Reticulum Contact Sites: A Perspective Review.DNA Cell Biol. 2020 Aug;39(8):1431-1443. doi: 10.1089/dna.2020.5614. Epub 2020 Jun 26. DNA Cell Biol. 2020. PMID: 32598172 Review.
-
Sensing endoplasmic reticulum stress by protein kinase RNA-like endoplasmic reticulum kinase promotes adaptive mitochondrial DNA biogenesis and cell survival via heme oxygenase-1/carbon monoxide activity.FASEB J. 2012 Jun;26(6):2558-68. doi: 10.1096/fj.11-199604. Epub 2012 Mar 5. FASEB J. 2012. PMID: 22391129
-
Dysfunctional mitochondria as critical players in the inflammation of autoimmune diseases: Potential role in Sjögren's syndrome.Autoimmun Rev. 2021 Aug;20(8):102867. doi: 10.1016/j.autrev.2021.102867. Epub 2021 Jun 9. Autoimmun Rev. 2021. PMID: 34118452 Review.
Cited by
-
Connection Between HIV and Mitochondria in Cardiovascular Disease and Implications for Treatments.Circ Res. 2024 May 24;134(11):1581-1606. doi: 10.1161/CIRCRESAHA.124.324296. Epub 2024 May 23. Circ Res. 2024. PMID: 38781302 Free PMC article. Review.
-
Modifications of Mitochondrial Network Morphology Affect the MAVS-Dependent Immune Response in L929 Murine Fibroblasts during Ectromelia Virus Infection.Pathogens. 2024 Aug 23;13(9):717. doi: 10.3390/pathogens13090717. Pathogens. 2024. PMID: 39338909 Free PMC article.
-
ER as master regulator of membrane trafficking and organelle function.J Cell Biol. 2022 Oct 3;221(10):e202205135. doi: 10.1083/jcb.202205135. Epub 2022 Sep 15. J Cell Biol. 2022. PMID: 36108241 Free PMC article. Review.
-
Redox regulation of UPR signalling and mitochondrial ER contact sites.Cell Mol Life Sci. 2024 Jun 7;81(1):250. doi: 10.1007/s00018-024-05286-0. Cell Mol Life Sci. 2024. PMID: 38847861 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

