Roles of MicroRNAs in Disease Biology
- PMID: 37179717
- PMCID: PMC10169270
- DOI: 10.31662/jmaj.2023-0009
Roles of MicroRNAs in Disease Biology
Abstract
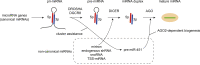
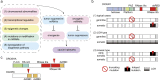
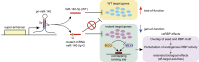
Gene regulation by microRNAs (miRNAs) plays important roles in development, physiology, and disease. miRNAs are an abundant class of noncoding RNAs that are generated through multistep biosynthetic pathways and typically repress gene expression through target destabilization and translational inhibition. Complex interactions between miRNAs and target mRNAs are associated with characteristic molecular mechanisms, including miRNA cotargeting, target-directed miRNA degradation, and crosstalk with various RNA-binding proteins. Consistent with the broad influence on cellular function, miRNA deregulation is commonly observed in various diseases, particularly cancer, with both tumor-suppressive and oncogenic roles. Mutations in the miRNA biosynthetic pathway and several miRNA genes have been linked to diverse types of cancer and a subset of genetic diseases, respectively. Additionally, super-enhancers play important roles in the regulation of cell type-specific and disease-associated miRNAs. This review summarizes the molecular features of miRNA biogenesis and target regulation along with the roles of miRNAs in disease biology, with recent examples expanding the pathophysiological roles of miRNAs.
Keywords: biogenesis; cancer; genetic disease; microRNA; target regulation.
Copyright © Japan Medical Association.
Conflict of interest statement
None
Figures

Similar articles
-
Systems and Synthetic microRNA Biology: From Biogenesis to Disease Pathogenesis.Int J Mol Sci. 2019 Dec 24;21(1):132. doi: 10.3390/ijms21010132. Int J Mol Sci. 2019. PMID: 31878193 Free PMC article. Review.
-
MicroRNAs: non coding pleiotropic factors in development, cancer prevention and treatment.Microrna. 2013;2(2):81. doi: 10.2174/2211536611302020001. Microrna. 2013. PMID: 25070777
-
miRNA and cancer; computational and experimental approaches.Curr Pharm Biotechnol. 2014;15(5):429. doi: 10.2174/138920101505140828161335. Curr Pharm Biotechnol. 2014. PMID: 25189575
-
The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression.Genome Biol. 2007;8(8):R166. doi: 10.1186/gb-2007-8-8-r166. Genome Biol. 2007. PMID: 17697356 Free PMC article.
-
MicroRNAs: molecular features and role in cancer.Front Biosci (Landmark Ed). 2012 Jun 1;17(7):2508-40. doi: 10.2741/4068. Front Biosci (Landmark Ed). 2012. PMID: 22652795 Free PMC article. Review.
Cited by
-
Post-transcriptional regulation of myogenic transcription factors during muscle development and pathogenesis.J Muscle Res Cell Motil. 2024 Mar;45(1):21-39. doi: 10.1007/s10974-023-09663-3. Epub 2024 Jan 11. J Muscle Res Cell Motil. 2024. PMID: 38206489 Review.
-
A Literature Review and Meta-Analysis on the Potential Use of miR-150 as a Novel Biomarker in the Detection and Progression of Multiple Sclerosis.J Pers Med. 2024 Jul 31;14(8):815. doi: 10.3390/jpm14080815. J Pers Med. 2024. PMID: 39202006 Free PMC article. Review.
-
Potential Diagnostic Utility of microRNAs in Gastrointestinal Cancers.Cancer Manag Res. 2023 Aug 22;15:863-871. doi: 10.2147/CMAR.S421928. eCollection 2023. Cancer Manag Res. 2023. PMID: 37636029 Free PMC article. Review.
-
Abnormal expressions of PURPL, miR-363-3p and ADAM10 predicted poor prognosis for patients with ovarian serous cystadenocarcinoma.J Cancer. 2023 Sep 11;14(15):2908-2918. doi: 10.7150/jca.87405. eCollection 2023. J Cancer. 2023. PMID: 37781085 Free PMC article.
-
Granulosa Cells-Related MicroRNAs in Ovarian Diseases: Mechanism, Facts and Perspectives.Reprod Sci. 2024 Dec;31(12):3635-3650. doi: 10.1007/s43032-024-01523-w. Epub 2024 Apr 9. Reprod Sci. 2024. PMID: 38594585 Review.
References
-
- Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5-20. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
