Validation of reference genes for the normalization of the RT-qPCR in peripheral blood mononuclear cells of septic patients
- PMID: 37089378
- PMCID: PMC10119759
- DOI: 10.1016/j.heliyon.2023.e15269
Validation of reference genes for the normalization of the RT-qPCR in peripheral blood mononuclear cells of septic patients
Abstract
Objective: To screen and validate reference genes suitable for gene mRNA expression study in peripheral blood mononuclear cells (PBMCs) between septic patients and healthy controls (HC).
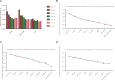
Methods: Total RNA in PBMCs was extracted and RT-qPCR was used to determine the mRNA expression profiles of 9 candidate genes, including ACTB, B2M, GAPDH, GUSB, HPRT1, PGK1, RPL13A, SDHA and YWHAZ. The genes expression stabilities were assessed by both geNorm and NormFinder software.
Results: YWHAZ was the most stable gene among the 9 candidate genes evaluated by both geNorm and NormFinder in mixed and sepsis groups. The most stable gene combination in mixed group analyzed by geNorm was the combination of GAPDH, PKG1 and YWHAZ, while that in sepsis group was the combination of ACTB, PKG1 and YWHAZ.
Conclusion: Our first systematic analysis of the reference genes in PBMC of septic patients suggested YWHAZ was the best candidate. The combination of ACTB, PKG1 and YWHAZ could improve RT-qPCR accuracy in septic patients. Our results identified the most stable reference genes to standardize RT-qPCR of sepsis patients, which can serve as a useful tool for gene function exploration in the future.
Keywords: RT-qPCR; Reference gene; Sepsis.
©2023PublishedbyElsevierLtd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Identification and validation of reference genes for RT-qPCR analysis in fetal rat pancreas.Reprod Toxicol. 2021 Oct;105:211-220. doi: 10.1016/j.reprotox.2021.09.009. Epub 2021 Sep 16. Reprod Toxicol. 2021. PMID: 34537367
-
Evaluation of suitable reference genes for gene expression studies in porcine PBMCs in response to LPS and LTA.BMC Res Notes. 2013 Feb 8;6:56. doi: 10.1186/1756-0500-6-56. BMC Res Notes. 2013. PMID: 23394600 Free PMC article.
-
Identification of optimal endogenous reference RNAs for RT-qPCR normalization in hindgut of rat models with anorectal malformations.PeerJ. 2019 Apr 23;7:e6829. doi: 10.7717/peerj.6829. eCollection 2019. PeerJ. 2019. PMID: 31065464 Free PMC article.
-
Evaluation of reference genes for human chondrocytes cultured in several different thermal environments.Int J Hyperthermia. 2014 May;30(3):210-6. doi: 10.3109/02656736.2014.906048. Int J Hyperthermia. 2014. PMID: 24773042
-
Time- and Region-Specific Selection of Reference Genes in the Rat Brain in the Lithium-Pilocarpine Model of Acquired Temporal Lobe Epilepsy.Biomedicines. 2024 May 16;12(5):1100. doi: 10.3390/biomedicines12051100. Biomedicines. 2024. PMID: 38791067 Free PMC article.
Cited by
-
Plac8-ERK pathway modulation of monocyte function in sepsis.Cell Death Discov. 2024 Jul 3;10(1):308. doi: 10.1038/s41420-024-02012-4. Cell Death Discov. 2024. PMID: 38961068 Free PMC article.
-
Identification and validation of immunity- and disulfidptosis-related genes signature for predicting prognosis in ovarian cancer.Heliyon. 2024 Jun 7;10(12):e32273. doi: 10.1016/j.heliyon.2024.e32273. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38952356 Free PMC article.
References
-
- Bustin S.A., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. - PubMed
-
- Chousterman B.G., Swirski F.K., Weber G.F. Cytokine storm and sepsis disease pathogenesis. Semin. Immunopathol. 2017;39(5):517–528. - PubMed
-
- Kaukonen K.M., et al. Systemic inflammatory response syndrome criteria in defining severe sepsis. N. Engl. J. Med. 2015;372(17):1629–1638. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous