Targeted Protein Degradation through E2 Recruitment
- PMID: 36940189
- PMCID: PMC10127205
- DOI: 10.1021/acschembio.3c00040
Targeted Protein Degradation through E2 Recruitment
Abstract
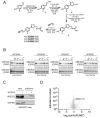
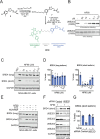
Targeted protein degradation (TPD) with proteolysis targeting chimeras (PROTACs), heterobifunctional compounds consisting of protein targeting ligands linked to recruiters of E3 ubiquitin ligases, has arisen as a powerful therapeutic modality to induce the proximity of target proteins with E3 ligases to ubiquitinate and degrade specific proteins in cells. Thus far, PROTACs have primarily exploited the recruitment of E3 ubiquitin ligases or their substrate adapter proteins but have not exploited the recruitment of more core components of the ubiquitin-proteasome system (UPS). In this study, we used covalent chemoproteomic approaches to discover a covalent recruiter against the E2 ubiquitin conjugating enzyme UBE2D─EN67─that targets an allosteric cysteine, C111, without affecting the enzymatic activity of the protein. We demonstrated that this UBE2D recruiter could be used in heterobifunctional degraders to degrade neo-substrate targets in a UBE2D-dependent manner, including BRD4 and the androgen receptor. Overall, our data highlight the potential for the recruitment of core components of the UPS machinery, such as E2 ubiquitin conjugating enzymes, for TPD, and underscore the utility of covalent chemoproteomic strategies for identifying novel recruiters for additional components of the UPS.
Conflict of interest statement
The authors declare the following competing financial interest(s): JAT, JMK, DD, MJH, MS are employees of Novartis Institutes for BioMedical Research. This study was funded by the Novartis Institutes for BioMedical Research and the Novartis-Berkeley Translational Chemical Biology Institute. DKN is a co-founder, shareholder, and scientific advisory board member for Frontier Medicines and Vicinitas Therapeutics. DKN is a member of the board of directors for Vicinitas Therapeutics. DKN is also on the scientific advisory board of The Mark Foundation for Cancer Research, MD Anderson Cancer Center, Photys Therapeutics, Apertor Pharmaceuticals, Oerth Bio, and Chordia Therapeutics. DKN is also an Investment Advisory Board member for Droia Ventures and a16z.
Figures


Similar articles
-
Targeted Protein Degradation through Recruitment of the CUL4 Complex Adaptor Protein DDB1.ACS Chem Biol. 2024 Jan 19;19(1):58-68. doi: 10.1021/acschembio.3c00487. Epub 2024 Jan 8. ACS Chem Biol. 2024. PMID: 38192078 Free PMC article.
-
Ligandability of E3 Ligases for Targeted Protein Degradation Applications.Biochemistry. 2023 Feb 7;62(3):588-600. doi: 10.1021/acs.biochem.1c00464. Epub 2021 Sep 2. Biochemistry. 2023. PMID: 34473924 Free PMC article. Review.
-
Exploiting the Cullin E3 Ligase Adaptor Protein SKP1 for Targeted Protein Degradation.ACS Chem Biol. 2024 Feb 16;19(2):442-450. doi: 10.1021/acschembio.3c00642. Epub 2024 Feb 2. ACS Chem Biol. 2024. PMID: 38305738 Free PMC article.
-
Targeted Protein Degradation through Recruitment of the CUL4A Complex Adaptor Protein DDB1.bioRxiv [Preprint]. 2023 Aug 12:2023.08.11.553046. doi: 10.1101/2023.08.11.553046. bioRxiv. 2023. Update in: ACS Chem Biol. 2024 Jan 19;19(1):58-68. doi: 10.1021/acschembio.3c00487. PMID: 37614621 Free PMC article. Updated. Preprint.
-
Ubiquitin proteasome system in immune regulation and therapeutics.Curr Opin Pharmacol. 2022 Dec;67:102310. doi: 10.1016/j.coph.2022.102310. Epub 2022 Oct 23. Curr Opin Pharmacol. 2022. PMID: 36288660 Free PMC article. Review.
Cited by
-
Insight into Recent Advances in Degrading Androgen Receptor for Castration-Resistant Prostate Cancer.Cancers (Basel). 2024 Feb 4;16(3):663. doi: 10.3390/cancers16030663. Cancers (Basel). 2024. PMID: 38339414 Free PMC article. Review.
-
Targeted protein degradation using chimeric human E2 ubiquitin-conjugating enzymes.Commun Biol. 2024 Sep 19;7(1):1179. doi: 10.1038/s42003-024-06803-4. Commun Biol. 2024. PMID: 39300128 Free PMC article.
-
The value of protein allostery in rational anticancer drug design: an update.Expert Opin Drug Discov. 2024 Sep;19(9):1071-1085. doi: 10.1080/17460441.2024.2384467. Epub 2024 Jul 28. Expert Opin Drug Discov. 2024. PMID: 39068599 Review.
-
DCAF16-Based Covalent Handle for the Rational Design of Monovalent Degraders.ACS Cent Sci. 2024 May 17;10(7):1318-1331. doi: 10.1021/acscentsci.4c00286. eCollection 2024 Jul 24. ACS Cent Sci. 2024. PMID: 39071058 Free PMC article.
-
Targeted protein degradation: from mechanisms to clinic.Nat Rev Mol Cell Biol. 2024 Sep;25(9):740-757. doi: 10.1038/s41580-024-00729-9. Epub 2024 Apr 29. Nat Rev Mol Cell Biol. 2024. PMID: 38684868 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

