Intestinal stem cell aging at single-cell resolution: Transcriptional perturbations alter cell developmental trajectory reversed by gerotherapeutics
- PMID: 36864750
- PMCID: PMC10186593
- DOI: 10.1111/acel.13802
Intestinal stem cell aging at single-cell resolution: Transcriptional perturbations alter cell developmental trajectory reversed by gerotherapeutics
Abstract
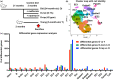
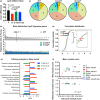
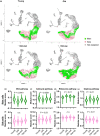
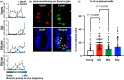
The intestinal epithelium consists of cells derived from continuously cycling Lgr5hi intestinal stem cells (Lgr5hi ISCs) that mature developmentally in an ordered fashion as the cells progress along the crypt-luminal axis. Perturbed function of Lgr5hi ISCs with aging is documented, but the consequent impact on overall mucosal homeostasis has not been defined. Using single-cell RNA sequencing, the progressive maturation of progeny was dissected in the mouse intestine, which revealed that transcriptional reprogramming with aging in Lgr5hi ISCs retarded the maturation of cells in their progression along the crypt-luminal axis. Importantly, treatment with metformin or rapamycin at a late stage of mouse lifespan reversed the effects of aging on the function of Lgr5hi ISCs and subsequent maturation of progenitors. The effects of metformin and rapamycin overlapped in reversing changes of transcriptional profiles but were also complementary, with metformin more efficient than rapamycin in correcting the developmental trajectory. Therefore, our data identify novel effects of aging on stem cells and the maturation of their daughter cells contributing to the decline of epithelial regeneration and the correction by geroprotectors.
Keywords: gerotherapeutics; intestinal stem cell aging; intestinal stem cells; metformin; rapamycin; single-cell RNA sequencing.
© 2023 The Authors. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
All authors state that there is no conflict of interest.
Figures

Similar articles
-
Fall of PARP3 restrains Lgr5+ intestinal stem cells proliferation and mucosal renovation in intestinal aging.Mech Ageing Dev. 2023 Apr;211:111796. doi: 10.1016/j.mad.2023.111796. Epub 2023 Mar 2. Mech Ageing Dev. 2023. PMID: 36870456
-
Distinct populations of embryonic epithelial progenitors generate Lgr5+ intestinal stem cells.Dev Biol. 2017 Dec 15;432(2):258-264. doi: 10.1016/j.ydbio.2017.10.012. Epub 2017 Oct 13. Dev Biol. 2017. PMID: 29037931
-
A constant pool of Lgr5+ intestinal stem cells is required for intestinal homeostasis.Cell Rep. 2021 Jan 26;34(4):108633. doi: 10.1016/j.celrep.2020.108633. Cell Rep. 2021. PMID: 33503423
-
Molecular regulation mechanism of intestinal stem cells in mucosal injury and repair in ulcerative colitis.World J Gastroenterol. 2023 Apr 28;29(16):2380-2396. doi: 10.3748/wjg.v29.i16.2380. World J Gastroenterol. 2023. PMID: 37179583 Free PMC article. Review.
-
Vitamin D and the nutritional environment in functions of intestinal stem cells: Implications for tumorigenesis and prevention.J Steroid Biochem Mol Biol. 2020 Apr;198:105556. doi: 10.1016/j.jsbmb.2019.105556. Epub 2019 Nov 26. J Steroid Biochem Mol Biol. 2020. PMID: 31783155 Free PMC article. Review.
Cited by
-
Dynamic Intestinal Stem Cell Plasticity and Lineage Remodeling by a Nutritional Environment Relevant to Human Risk for Tumorigenesis.Mol Cancer Res. 2023 Aug 1;21(8):808-824. doi: 10.1158/1541-7786.MCR-22-1000. Mol Cancer Res. 2023. PMID: 37097719 Free PMC article.
-
A cellular identity crisis? Plasticity changes during aging and rejuvenation.Genes Dev. 2024 Oct 16;38(17-20):823-842. doi: 10.1101/gad.351728.124. Genes Dev. 2024. PMID: 39293862 Free PMC article. Review.
-
Gut aging: A wane from the normal to repercussion and gerotherapeutic strategies.Heliyon. 2024 Sep 12;10(19):e37883. doi: 10.1016/j.heliyon.2024.e37883. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39381110 Free PMC article. Review.
-
Dietary and metabolic effects on intestinal stem cells in health and disease.Nat Rev Gastroenterol Hepatol. 2025 Jan;22(1):23-38. doi: 10.1038/s41575-024-00980-7. Epub 2024 Oct 2. Nat Rev Gastroenterol Hepatol. 2025. PMID: 39358589 Review.
-
The cell rejuvenation atlas: leveraging network biology to identify master regulators of rejuvenation strategies.Aging (Albany NY). 2024 Sep 9;16(17):12168-12190. doi: 10.18632/aging.206105. Epub 2024 Sep 9. Aging (Albany NY). 2024. PMID: 39264584 Free PMC article.
References
-
- Barker, N. , van Es, J. H. , Kuipers, J. , Kujala, P. , van den Born, M. , Cozijnsen, M. , Haegebarth, A. , Korving, J. , Begthel, H. , Peters, P. J. , & Clevers, H. (2007). Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 449(7165), 1003–1007. 10.1038/nature06196 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

