Differential expression of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) in Alzheimer's disease and HIV-1 associated neurocognitive disorders
- PMID: 36841839
- PMCID: PMC9968324
- DOI: 10.1038/s41598-022-27276-7
Differential expression of interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) in Alzheimer's disease and HIV-1 associated neurocognitive disorders
Abstract
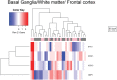
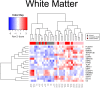
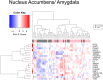
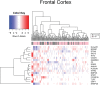
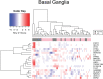
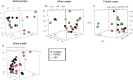
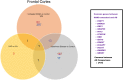
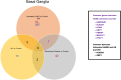
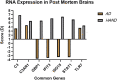
The United Nations projects that one in every six people will be over the age of 65 by the year 2050. With a rapidly aging population, the risk of Alzheimer's disease (AD) becomes a major concern. AD is a multifactorial disease that involves neurodegeneration in the brain with mild dementia and deficits in memory and other cognitive domains. Additionally, it has been established that individuals with Human Immunodeficiency Virus-1 (HIV-1) experience a 5 to 10-year accelerated aging and an increased risk of developing HIV-associated neurocognitive disorders (HAND). Despite a significant amount of clinical evidence pointing towards a potential overlap between neuropathogenic processes in HAND and AD, the underlying epigenetic link between these two diseases is mostly unknown. This study is focused on identifying differentially expressed genes observed in both AD and HAND using linear regression models and a more robust significance analysis of microarray. The results established that the dysregulated type 1 and 2 interferon pathways observed in both AD and HAND contribute to the similar pathologies of these diseases within the brain. The current study identifies the important roles of interferon pathways in AD and HAND, a relationship that may be useful for earlier detection in the future.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The current understanding of overlap between characteristics of HIV-associated neurocognitive disorders and Alzheimer's disease.J Neurovirol. 2019 Oct;25(5):661-672. doi: 10.1007/s13365-018-0702-9. Epub 2019 Jan 22. J Neurovirol. 2019. PMID: 30671777 Free PMC article. Review.
-
Does HIV infection contribute to increased beta-amyloid synthesis and plaque formation leading to neurodegeneration and Alzheimer's disease?J Neurovirol. 2019 Oct;25(5):634-647. doi: 10.1007/s13365-019-00732-3. Epub 2019 Mar 13. J Neurovirol. 2019. PMID: 30868421 Review.
-
Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer's disease.J Neurovirol. 2019 Oct;25(5):702-709. doi: 10.1007/s13365-018-0695-4. Epub 2019 Jan 4. J Neurovirol. 2019. PMID: 30610738 Free PMC article. Review.
-
Systems analysis of human brain gene expression: mechanisms for HIV-associated neurocognitive impairment and common pathways with Alzheimer's disease.BMC Med Genomics. 2013 Feb 13;6:4. doi: 10.1186/1755-8794-6-4. BMC Med Genomics. 2013. PMID: 23406646 Free PMC article.
-
Brain and liver pathology, amyloid deposition, and interferon responses among older HIV-positive patients in the late HAART era.BMC Infect Dis. 2017 Feb 17;17(1):151. doi: 10.1186/s12879-017-2246-7. BMC Infect Dis. 2017. PMID: 28212619 Free PMC article.
Cited by
-
Neuropathogenic role of astrocyte-derived extracellular vesicles in HIV-associated neurocognitive disorders.J Extracell Vesicles. 2024 Apr;13(4):e12439. doi: 10.1002/jev2.12439. J Extracell Vesicles. 2024. PMID: 38647111 Free PMC article.
-
Navigating the Gene Co-Expression Network and Drug Repurposing Opportunities for Brain Disorders Associated with Neurocognitive Impairment.Brain Sci. 2023 Nov 7;13(11):1564. doi: 10.3390/brainsci13111564. Brain Sci. 2023. PMID: 38002524 Free PMC article.
-
Clinical and CSF single-cell profiling of post-COVID-19 cognitive impairment.Cell Rep Med. 2024 May 21;5(5):101561. doi: 10.1016/j.xcrm.2024.101561. Epub 2024 May 13. Cell Rep Med. 2024. PMID: 38744274 Free PMC article.
References
-
- Milanini B, Samboju V, Cobigo Y, Paul R, Javandel S, Hellmuth J, Allen I, Miller B, Valcour V. Longitudinal brain atrophy patterns and neuropsychological performance in older adults with HIV-associated neurocognitive disorder compared to early alzheimer's disease. Neurobiol. Aging. 2019;82:69–76. doi: 10.1016/j.neurobiolaging.2019.07.006. - DOI - PMC - PubMed

