Quantitative analysis of sterol-modulated monomer-dimer equilibrium of the β1-adrenergic receptor by DEER spectroscopy
- PMID: 36745787
- PMCID: PMC9963004
- DOI: 10.1073/pnas.2221036120
Quantitative analysis of sterol-modulated monomer-dimer equilibrium of the β1-adrenergic receptor by DEER spectroscopy
Abstract
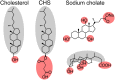
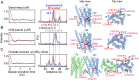
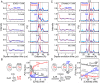
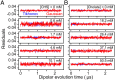
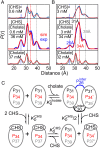
G protein-coupled receptors (GPCR) activate numerous intracellular signaling pathways. The oligomerization properties of GPCRs, and hence their cellular functions, may be modulated by various components within the cell membrane (such as the presence of cholesterol). Modulation may occur directly via specific interaction with the GPCR or indirectly by affecting the physical properties of the membrane. Here, we use pulsed Q-band double electron-electron resonance (DEER) spectroscopy to probe distances between R1 nitroxide spin labels attached to Cys163 and Cys344 of the β1-adrenergic receptor (β1AR) in n-dodecyl-β-D-maltoside micelles upon titration with two soluble cholesterol analogs, cholesteryl hemisuccinate (CHS) and sodium cholate. The former, like cholesterol, inserts itself into the lipid membrane, parallel to the phospholipid chains; the latter is aligned parallel to the surface of membranes. Global quantitative analysis of DEER echo curves upon titration of spin-labeled β1AR with CHS and sodium cholate reveal the following: CHS binds specifically to the β1AR monomer at a site close to the Cys163-R1 spin label with an equilibrium dissociation constant [Formula: see text] ~1.4 ± 0.4 mM. While no direct binding of sodium cholate to the β1AR receptor was observed by DEER, sodium cholate induces specific β1AR dimerization ([Formula: see text] ~35 ± 6 mM and a Hill coefficient n ~ 2.5 ± 0.4) with intersubunit contacts between transmembrane helices 1 and 2 and helix 8. Analysis of the DEER data obtained upon the addition of CHS to the β1AR dimer in the presence of excess cholate results in dimer dissociation with species occupancies as predicted from the individual KD values.
Keywords: G protein-coupled receptor; cholesterol; dimerization; electron paramagnetic resonance; spin-labeling.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Deconvoluting Monomer- and Dimer-Specific Distance Distributions between Spin Labels in a Monomer/Dimer Mixture Using T1-Edited DEER EPR Spectroscopy.J Am Chem Soc. 2024 Jul 3;146(26):17964-17973. doi: 10.1021/jacs.4c03916. Epub 2024 Jun 18. J Am Chem Soc. 2024. PMID: 38888555 Free PMC article.
-
DEER Analysis of GPCR Conformational Heterogeneity.Biomolecules. 2021 May 22;11(6):778. doi: 10.3390/biom11060778. Biomolecules. 2021. PMID: 34067265 Free PMC article. Review.
-
EPR Techniques to Probe Insertion and Conformation of Spin-Labeled Proteins in Lipid Bilayers.Methods Mol Biol. 2019;2003:493-528. doi: 10.1007/978-1-4939-9512-7_21. Methods Mol Biol. 2019. PMID: 31218631
-
Quantitative Resolution of Monomer-Dimer Populations by Inversion Modulated DEER EPR Spectroscopy.Chemphyschem. 2016 Oct 5;17(19):2987-2991. doi: 10.1002/cphc.201600726. Epub 2016 Aug 2. Chemphyschem. 2016. PMID: 27442455 Free PMC article.
-
Time-resolved electron spin resonance studies of spin-labelled lipids in membranes.Chem Phys Lipids. 2006 Jun;141(1-2):142-57. doi: 10.1016/j.chemphyslip.2006.02.009. Epub 2006 Mar 13. Chem Phys Lipids. 2006. PMID: 16564516 Review.
Cited by
-
Deconvoluting Monomer- and Dimer-Specific Distance Distributions between Spin Labels in a Monomer/Dimer Mixture Using T1-Edited DEER EPR Spectroscopy.J Am Chem Soc. 2024 Jul 3;146(26):17964-17973. doi: 10.1021/jacs.4c03916. Epub 2024 Jun 18. J Am Chem Soc. 2024. PMID: 38888555 Free PMC article.
-
Xplor-NIH: Better parameters and protocols for NMR protein structure determination.Protein Sci. 2024 Apr;33(4):e4922. doi: 10.1002/pro.4922. Protein Sci. 2024. PMID: 38501482
-
G Protein-Coupled Receptor Dimerization-What Next?Int J Mol Sci. 2024 Mar 7;25(6):3089. doi: 10.3390/ijms25063089. Int J Mol Sci. 2024. PMID: 38542063 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

