REEP1 Preserves Motor Function in SOD1G93A Mice by Improving Mitochondrial Function via Interaction with NDUFA4
- PMID: 36520405
- PMCID: PMC10264344
- DOI: 10.1007/s12264-022-00995-7
REEP1 Preserves Motor Function in SOD1G93A Mice by Improving Mitochondrial Function via Interaction with NDUFA4
Abstract
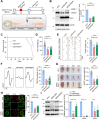
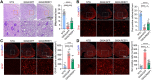
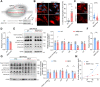
A decline in the activities of oxidative phosphorylation (OXPHOS) complexes has been consistently reported in amyotrophic lateral sclerosis (ALS) patients and animal models of ALS, although the underlying molecular mechanisms are still elusive. Here, we report that receptor expression enhancing protein 1 (REEP1) acts as an important regulator of complex IV assembly, which is pivotal to preserving motor neurons in SOD1G93A mice. We found the expression of REEP1 was greatly reduced in transgenic SOD1G93A mice with ALS. Moreover, forced expression of REEP1 in the spinal cord extended the lifespan, decelerated symptom progression, and improved the motor performance of SOD1G93A mice. The neuromuscular synaptic loss, gliosis, and even motor neuron loss in SOD1G93A mice were alleviated by increased REEP1 through augmentation of mitochondrial function. Mechanistically, REEP1 associates with NDUFA4, and plays an important role in preserving the integrity of mitochondrial complex IV. Our findings offer insights into the pathogenic mechanism of REEP1 deficiency in neurodegenerative diseases and suggest a new therapeutic target for ALS.
Keywords: Amyotrophic lateral sclerosis; Complex IV assembly; Mitochondria; NDUFA4; REEP1.
© 2022. Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
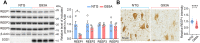

Similar articles
-
R13 preserves motor performance in SOD1G93A mice by improving mitochondrial function.Theranostics. 2021 May 24;11(15):7294-7307. doi: 10.7150/thno.56070. eCollection 2021. Theranostics. 2021. PMID: 34158851 Free PMC article.
-
Modulation of astrocytic mitochondrial function by dichloroacetate improves survival and motor performance in inherited amyotrophic lateral sclerosis.PLoS One. 2012;7(4):e34776. doi: 10.1371/journal.pone.0034776. Epub 2012 Apr 3. PLoS One. 2012. PMID: 22509356 Free PMC article.
-
Knocking down metabotropic glutamate receptor 1 improves survival and disease progression in the SOD1(G93A) mouse model of amyotrophic lateral sclerosis.Neurobiol Dis. 2014 Apr;64:48-59. doi: 10.1016/j.nbd.2013.11.006. Epub 2013 Dec 19. Neurobiol Dis. 2014. PMID: 24361555
-
Pharmacological inhibition of ALCAT1 mitigates amyotrophic lateral sclerosis by attenuating SOD1 protein aggregation.Mol Metab. 2022 Sep;63:101536. doi: 10.1016/j.molmet.2022.101536. Epub 2022 Jun 28. Mol Metab. 2022. PMID: 35772643 Free PMC article.
-
Novel behavioural characteristics of the superoxide dismutase 1 G93A (SOD1G93A ) mouse model of amyotrophic lateral sclerosis include sex-dependent phenotypes.Genes Brain Behav. 2020 Feb;19(2):e12604. doi: 10.1111/gbb.12604. Epub 2019 Sep 10. Genes Brain Behav. 2020. PMID: 31412164
Cited by
-
Axonopathy Underlying Amyotrophic Lateral Sclerosis: Unraveling Complex Pathways and Therapeutic Insights.Neurosci Bull. 2024 Nov;40(11):1789-1810. doi: 10.1007/s12264-024-01267-2. Epub 2024 Aug 4. Neurosci Bull. 2024. PMID: 39097850 Review.
-
Mitochondrial respiratory chain component NDUFA4: a promising therapeutic target for gastrointestinal cancer.Cancer Cell Int. 2024 Mar 5;24(1):97. doi: 10.1186/s12935-024-03283-8. Cancer Cell Int. 2024. PMID: 38443961 Free PMC article. Review.
-
Exploration of Potential Target Genes of miR-24-3p in Chicken Myoblasts by Transcriptome Sequencing Analysis.Genes (Basel). 2023 Sep 5;14(9):1764. doi: 10.3390/genes14091764. Genes (Basel). 2023. PMID: 37761904 Free PMC article.
-
Hypoxic Preconditioned Neural Stem Cell-Derived Extracellular Vesicles Contain Distinct Protein Cargo from Their Normal Counterparts.Curr Issues Mol Biol. 2023 Mar 1;45(3):1982-1997. doi: 10.3390/cimb45030127. Curr Issues Mol Biol. 2023. PMID: 36975497 Free PMC article.
-
Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes.Biomolecules. 2024 Mar 20;14(3):377. doi: 10.3390/biom14030377. Biomolecules. 2024. PMID: 38540795 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

