Loss of Drosophila Clueless differentially affects the mitochondrial proteome compared to loss of Sod2 and Pink1
- PMID: 36388112
- PMCID: PMC9644064
- DOI: 10.3389/fphys.2022.1004099
Loss of Drosophila Clueless differentially affects the mitochondrial proteome compared to loss of Sod2 and Pink1
Abstract
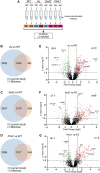
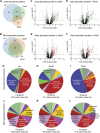
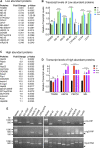
Mitochondria contain their own DNA, mitochondrial DNA, which encodes thirteen proteins. However, mitochondria require thousands of proteins encoded in the nucleus to carry out their many functions. Identifying the definitive mitochondrial proteome has been challenging as methods isolating mitochondrial proteins differ and different tissues and organisms may have specialized proteomes. Mitochondrial diseases arising from single gene mutations in nucleus encoded genes could affect the mitochondrial proteome, but deciphering which effects are due to loss of specific pathways or to accumulated general mitochondrial damage is difficult. To identify specific versus general effects, we have taken advantage of mutations in three Drosophila genes, clueless, Sod2, and Pink1, which are required for mitochondrial function through different pathways. We measured changes in each mutant's mitochondrial proteome using quantitative tandem mass tag mass spectrometry. Our analysis identified protein classes that are unique to each mutant and those shared between them, suggesting that some changes in the mitochondrial proteome are due to general mitochondrial damage whereas others are gene specific. For example, clueless mutants had the greatest number of less and more abundant mitochondrial proteins whereas loss of all three genes increased stress and metabolism proteins. This study is the first to directly compare in vivo steady state levels of mitochondrial proteins by examining loss of three pathways critical for mitochondrial function. These data could be useful to understand disease etiology, and how mutations in genes critical for mitochondrial function cause specific mitochondrial proteomic changes as opposed to changes due to generalized mitochondrial damage.
Keywords: Clueless; PINK1; SOD2; drosophila; mitochondria; mitochondrial proteome; respiratory chain complexes.
Copyright © 2022 Sen and Cox.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Clueless, a protein required for mitochondrial function, interacts with the PINK1-Parkin complex in Drosophila.Dis Model Mech. 2015 Jun;8(6):577-89. doi: 10.1242/dmm.019208. Epub 2015 Apr 20. Dis Model Mech. 2015. PMID: 26035866 Free PMC article.
-
The dysregulated Pink1- Drosophila mitochondrial proteome is partially corrected with exercise.Aging (Albany NY). 2021 Jun 1;13(11):14709-14728. doi: 10.18632/aging.203128. Epub 2021 Jun 1. Aging (Albany NY). 2021. PMID: 34074800 Free PMC article.
-
Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants.J Proteomics. 2014 Sep 23;109:400-16. doi: 10.1016/j.jprot.2014.07.005. Epub 2014 Jul 12. J Proteomics. 2014. PMID: 25026440
-
N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review.
-
PINK1-Parkin signaling in Parkinson's disease: Lessons from Drosophila.Neurosci Res. 2020 Oct;159:40-46. doi: 10.1016/j.neures.2020.01.016. Epub 2020 Feb 6. Neurosci Res. 2020. PMID: 32035987 Review.
Cited by
-
Drosophila Clueless ribonucleoprotein particles display novel dynamics that rely on the availability of functional protein and polysome equilibrium.bioRxiv [Preprint]. 2024 Aug 22:2024.08.21.609023. doi: 10.1101/2024.08.21.609023. bioRxiv. 2024. PMID: 39229069 Free PMC article. Preprint.
-
CLUH maintains functional mitochondria and translation in motoneuronal axons and prevents peripheral neuropathy.Sci Adv. 2024 May 31;10(22):eadn2050. doi: 10.1126/sciadv.adn2050. Epub 2024 May 29. Sci Adv. 2024. PMID: 38809982 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

