Niclosamide targets the dynamic progression of macrophages for the resolution of endometriosis in a mouse model
- PMID: 36369244
- PMCID: PMC9652344
- DOI: 10.1038/s42003-022-04211-0
Niclosamide targets the dynamic progression of macrophages for the resolution of endometriosis in a mouse model
Abstract
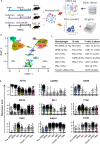
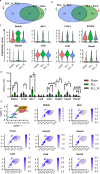
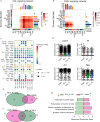
Due to the vital roles of macrophages in the pathogenesis of endometriosis, targeting macrophages could be a promising therapeutic direction. Here, we investigated the efficacy of niclosamide for the resolution of a perturbed microenvironment caused by dysregulated macrophages in a mouse model of endometriosis. Single-cell transcriptomic analysis revealed the heterogeneity of macrophages including three intermediate subtypes with sharing characteristics of traditional "small" or "large" peritoneal macrophages (SPMs and LPMs) in the peritoneal cavity. Endometriosis-like lesions (ELL) enhanced the differentiation of recruited macrophages, promoted the replenishment of resident LPMs, and increased the ablation of embryo-derived LPMs, which were stepwise suppressed by niclosamide. In addition, niclosamide restored intercellular communications between macrophages and B cells. Therefore, niclosamide rescued the perturbed microenvironment in endometriosis through its fine regulations on the dynamic progression of macrophages. Validation of similar macrophage pathogenesis in patients will further promote the clinical usage of niclosamide for endometriosis treatment.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Rediscovering peritoneal macrophages in a murine endometriosis model.Hum Reprod. 2017 Jan;32(1):94-102. doi: 10.1093/humrep/dew274. Epub 2016 Nov 5. Hum Reprod. 2017. PMID: 27816922
-
Efficacy of niclosamide on the intra-abdominal inflammatory environment in endometriosis.FASEB J. 2021 May;35(5):e21584. doi: 10.1096/fj.202002541RRR. FASEB J. 2021. PMID: 33860549 Free PMC article.
-
Niclosamide suppresses macrophage-induced inflammation in endometriosis†.Biol Reprod. 2020 Apr 24;102(5):1011-1019. doi: 10.1093/biolre/ioaa010. Biol Reprod. 2020. PMID: 31950153 Free PMC article.
-
Niclosamide: Beyond an antihelminthic drug.Cell Signal. 2018 Jan;41:89-96. doi: 10.1016/j.cellsig.2017.04.001. Epub 2017 Apr 4. Cell Signal. 2018. PMID: 28389414 Free PMC article. Review.
-
The Role of Peritoneal Macrophages in Endometriosis.Int J Mol Sci. 2021 Oct 6;22(19):10792. doi: 10.3390/ijms221910792. Int J Mol Sci. 2021. PMID: 34639133 Free PMC article. Review.
Cited by
-
New Potential Pharmacological Options for Endometriosis Associated Pain.Int J Mol Sci. 2024 Jun 27;25(13):7068. doi: 10.3390/ijms25137068. Int J Mol Sci. 2024. PMID: 39000175 Free PMC article. Review.
-
The involvement of peritoneal GATA6+ macrophages in the pathogenesis of endometriosis.Front Immunol. 2024 Aug 12;15:1396000. doi: 10.3389/fimmu.2024.1396000. eCollection 2024. Front Immunol. 2024. PMID: 39192982 Free PMC article.
-
MMP-9 as a clinical marker for endometriosis: a meta-analysis and bioinformatics analysis.Front Endocrinol (Lausanne). 2024 Oct 31;15:1475531. doi: 10.3389/fendo.2024.1475531. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39544239 Free PMC article.
-
High-fat diets promote peritoneal inflammation and augment endometriosis-associated abdominal hyperalgesia.bioRxiv [Preprint]. 2023 Nov 13:2023.11.09.566474. doi: 10.1101/2023.11.09.566474. bioRxiv. 2023. Update in: Front Endocrinol (Lausanne). 2024 Mar 15;15:1336496. doi: 10.3389/fendo.2024.1336496 PMID: 38014254 Free PMC article. Updated. Preprint.
-
High-fat diets promote peritoneal inflammation and augment endometriosis-associated abdominal hyperalgesia.Front Endocrinol (Lausanne). 2024 Mar 15;15:1336496. doi: 10.3389/fendo.2024.1336496. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 38559689 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

