SARS-CoV-2 spike opening dynamics and energetics reveal the individual roles of glycans and their collective impact
- PMID: 36329138
- PMCID: PMC9631587
- DOI: 10.1038/s42003-022-04138-6
SARS-CoV-2 spike opening dynamics and energetics reveal the individual roles of glycans and their collective impact
Abstract
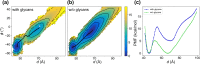
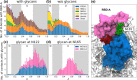
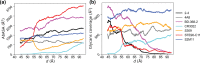
The trimeric spike (S) glycoprotein, which protrudes from the SARS-CoV-2 viral envelope, binds to human ACE2, initiated by at least one protomer's receptor binding domain (RBD) switching from a "down" (closed) to an "up" (open) state. Here, we used large-scale molecular dynamics simulations and two-dimensional replica exchange umbrella sampling calculations with more than a thousand windows and an aggregate total of 160 μs of simulation to investigate this transition with and without glycans. We find that the glycosylated spike has a higher barrier to opening and also energetically favors the down state over the up state. Analysis of the S-protein opening pathway reveals that glycans at N165 and N122 interfere with hydrogen bonds between the RBD and the N-terminal domain in the up state, while glycans at N165 and N343 can stabilize both the down and up states. Finally, we estimate how epitope exposure for several known antibodies changes along the opening path. We find that the BD-368-2 antibody's epitope is continuously exposed, explaining its high efficacy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
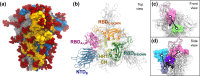


Similar articles
-
All-atom simulations of the trimeric spike protein of SARS-CoV-2 in aqueous medium: Nature of interactions, conformational stability and free energy diagrams for conformational transition of the protein.J Comput Chem. 2023 Jun 30;44(17):1560-1577. doi: 10.1002/jcc.27108. Epub 2023 Mar 31. J Comput Chem. 2023. PMID: 37000187
-
Static all-atom energetic mappings of the SARS-Cov-2 spike protein and dynamic stability analysis of "Up" versus "Down" protomer states.PLoS One. 2020 Nov 10;15(11):e0241168. doi: 10.1371/journal.pone.0241168. eCollection 2020. PLoS One. 2020. PMID: 33170884 Free PMC article.
-
Energetics of Spike Protein Opening of SARS-CoV-1 and SARS-CoV-2 and Its Variants of Concern: Implications in Host Receptor Scanning and Transmission.Biochemistry. 2022 Oct 18;61(20):2188-2197. doi: 10.1021/acs.biochem.2c00301. Epub 2022 Sep 27. Biochemistry. 2022. PMID: 36166360
-
Many Roles of Carbohydrates: A Computational Spotlight on the Coronavirus S Protein Binding.ACS Appl Bio Mater. 2024 Feb 19;7(2):646-656. doi: 10.1021/acsabm.2c01064. Epub 2023 Mar 22. ACS Appl Bio Mater. 2024. PMID: 36947738 Free PMC article. Review.
-
Structural basis of severe acute respiratory syndrome coronavirus 2 infection.Curr Opin HIV AIDS. 2021 Jan;16(1):74-81. doi: 10.1097/COH.0000000000000658. Curr Opin HIV AIDS. 2021. PMID: 33186231 Review.
Cited by
-
Conserved role of spike S2 domain N-glycosylation across beta-coronavirus family.bioRxiv [Preprint]. 2024 Sep 5:2024.09.05.611372. doi: 10.1101/2024.09.05.611372. bioRxiv. 2024. PMID: 39282346 Free PMC article. Preprint.
-
Water-glycan interactions drive the SARS-CoV-2 spike dynamics: insights into glycan-gate control and camouflage mechanisms.Chem Sci. 2024 Aug 23;15(35):14177-87. doi: 10.1039/d4sc04364b. Online ahead of print. Chem Sci. 2024. PMID: 39220162 Free PMC article.
-
The effects of amino acid substitution of spike protein and genomic recombination on the evolution of SARS-CoV-2.Front Microbiol. 2023 Jul 25;14:1228128. doi: 10.3389/fmicb.2023.1228128. eCollection 2023. Front Microbiol. 2023. PMID: 37560529 Free PMC article. Review.
-
The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.PLoS Pathog. 2022 Jul 11;18(7):e1010631. doi: 10.1371/journal.ppat.1010631. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35816514 Free PMC article.
-
N-Glycome Profile of the Spike Protein S1: Systemic and Comparative Analysis from Eleven Variants of SARS-CoV-2.Biomolecules. 2023 Sep 20;13(9):1421. doi: 10.3390/biom13091421. Biomolecules. 2023. PMID: 37759821 Free PMC article.
References
-
- Owen DR, et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374:1586–1593. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

