Tamarix hispida NAC Transcription Factor ThNAC4 Confers Salt and Drought Stress Tolerance to Transgenic Tamarix and Arabidopsis
- PMID: 36235512
- PMCID: PMC9570625
- DOI: 10.3390/plants11192647
Tamarix hispida NAC Transcription Factor ThNAC4 Confers Salt and Drought Stress Tolerance to Transgenic Tamarix and Arabidopsis
Abstract
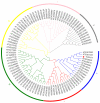
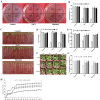
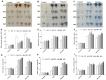
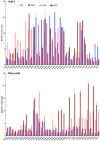
Salt and drought are considered two major abiotic stresses that have a significant impact on plants. Plant NAC (NAM, ATAF1/2, and CUC2) transcription factors (TFs) have been shown to play vital roles in plant development and responses to various abiotic stresses. ThNAC4, a NAC gene from Tamarix hispida involved in salt and osmotic stress tolerance, was identified and characterized in this study. According to a phylogenetic study, ThNAC4 is a member of NAC subfamily II. Subcellular localization analysis showed that ThNAC4 is located in the nucleus, and transcriptional activation experiments demonstrated that ThNAC4 is a transcriptional activator. Transgenic Arabidopsis plants overexpressing ThNAC4 exhibited improved salt and osmotic tolerance, as demonstrated by improved physiological traits. ThNAC4-overexpressing and ThNAC4-silenced T. hispida plants were generated using the transient transformation method and selected for gain- and loss-of-function analysis. The results showed that overexpression of ThNAC4 in transgenic Tamarix and Arabidopsis plants increased the activities of antioxidant enzymes (SOD, POD, and GST) and osmoprotectant (proline and trehalose) contents under stress conditions. These findings suggest that ThNAC4 plays an important physiological role in plant abiotic stress tolerance by increasing ROS scavenging ability and improving osmotic potential.
Keywords: NAC transcription factor; ROS scavenging; Tamarix hispida; abiotic stress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
ThNAC13, a NAC Transcription Factor from Tamarix hispida, Confers Salt and Osmotic Stress Tolerance to Transgenic Tamarix and Arabidopsis.Front Plant Sci. 2017 Apr 26;8:635. doi: 10.3389/fpls.2017.00635. eCollection 2017. Front Plant Sci. 2017. PMID: 28491072 Free PMC article.
-
The NAC Protein from Tamarix hispida, ThNAC7, Confers Salt and Osmotic Stress Tolerance by Increasing Reactive Oxygen Species Scavenging Capability.Plants (Basel). 2019 Jul 12;8(7):221. doi: 10.3390/plants8070221. Plants (Basel). 2019. PMID: 31336966 Free PMC article.
-
ThASR3 confers salt and osmotic stress tolerances in transgenic Tamarix and Arabidopsis.BMC Plant Biol. 2022 Dec 14;22(1):586. doi: 10.1186/s12870-022-03942-w. BMC Plant Biol. 2022. PMID: 36517747 Free PMC article.
-
An ERF transcription factor from Tamarix hispida, ThCRF1, can adjust osmotic potential and reactive oxygen species scavenging capability to improve salt tolerance.Plant Sci. 2017 Dec;265:154-166. doi: 10.1016/j.plantsci.2017.10.006. Epub 2017 Oct 13. Plant Sci. 2017. PMID: 29223337
-
NAC transcription factors in plant abiotic stress responses.Biochim Biophys Acta. 2012 Feb;1819(2):97-103. doi: 10.1016/j.bbagrm.2011.10.005. Epub 2011 Oct 19. Biochim Biophys Acta. 2012. PMID: 22037288 Review.
Cited by
-
Picea wilsonii NAC31 and DREB2A Cooperatively Activate ERD1 to Modulate Drought Resistance in Transgenic Arabidopsis.Int J Mol Sci. 2024 Feb 7;25(4):2037. doi: 10.3390/ijms25042037. Int J Mol Sci. 2024. PMID: 38396714 Free PMC article.
-
Physiological and Transcriptional Responses of Apocynum venetum to Salt Stress at the Seed Germination Stage.Int J Mol Sci. 2023 Feb 11;24(4):3623. doi: 10.3390/ijms24043623. Int J Mol Sci. 2023. PMID: 36835035 Free PMC article.
-
A Transcription Factor SlNAC4 Gene of Suaeda liaotungensis Enhances Salt and Drought Tolerance through Regulating ABA Synthesis.Plants (Basel). 2023 Aug 15;12(16):2951. doi: 10.3390/plants12162951. Plants (Basel). 2023. PMID: 37631162 Free PMC article.
-
Genome-Wide Identification of NAC Family Genes in Oat and Functional Characterization of AsNAC109 in Abiotic Stress Tolerance.Plants (Basel). 2024 Apr 3;13(7):1017. doi: 10.3390/plants13071017. Plants (Basel). 2024. PMID: 38611546 Free PMC article.
-
Evolution of Duplicated Glutathione Metabolic Pathway in Gossypium hirsutum and Its Response to UV-B Stress.Ecol Evol. 2024 Nov 19;14(11):e70537. doi: 10.1002/ece3.70537. eCollection 2024 Nov. Ecol Evol. 2024. PMID: 39563703 Free PMC article.
References
-
- Manzoor M.A., Manzoor M.M., Li G., Abdullah M., Han W., Wenlong H., Shakoor A., Riaz M.W., Rehman S., Cai Y. Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear (Pyrus bretschneider i) BMC Plant Biol. 2021;21:413. doi: 10.1186/s12870-021-03191-3. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

