Gut metagenome-derived signature predicts hepatic decompensation and mortality in NAFLD-related cirrhosis
- PMID: 36164267
- PMCID: PMC9746351
- DOI: 10.1111/apt.17236
Gut metagenome-derived signature predicts hepatic decompensation and mortality in NAFLD-related cirrhosis
Abstract
Background: There are limited data on the diagnostic accuracy of gut microbial signatures for predicting hepatic decompensation in patients with cirrhosis.
Aims: To determine whether a stool metagenome-derived signature accurately detects hepatic decompensation and mortality risk in cirrhosis secondary to non-alcoholic fatty liver disease (NAFLD) METHODS: Shotgun metagenomic sequencing was performed on faecal samples collected at study entry from a prospective cohort of adults with NAFLD-related cirrhosis. A Random Forest machine learning algorithm was utilised to identify a metagenomic signature of decompensated cirrhosis (defined by ascites, hepatic encephalopathy or variceal haemorrhage) and subsequently validated in an external cohort. A Cox proportional hazards regression model was used to examine predictors of all-cause mortality.
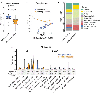
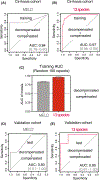
Results: In all, 25 adults with NAFLD-related cirrhosis (training cohort) were included. Among the 16 participants with decompensated cirrhosis, 33% had ascites, 56% had hepatic encephalopathy and 22% had experienced a variceal haemorrhage (not mutually exclusive). We identified a stool metagenomic signature comprising 13 discriminatory species that reliably distinguished decompensated NAFLD-related cirrhosis (diagnostic accuracy, 0.97, 95% confidence interval [CI] 0.96-0.99). Diagnostic accuracy of the 13-species signature remained high after adjustment for lactulose (area under the curve [AUC] 0.99) and rifaximin use (AUC 0.93). The discriminative ability of 13-species metagenomic signature was robust in an independent test cohort (AUC 0.95, 95% CI 0.81-1.00). The 13-species metagenomic signature (hazard ratio [HR] 1.54, 95% CI 1.10-2.15, p = 0.01) was a stronger predictor of mortality than the Model for End-Stage Liver Disease score (HR 1.25, 95% CI 1.03-1.53, p = 0.03).
Conclusions: This study provides evidence for a gut metagenome-derived signature with high diagnostic accuracy for hepatic decompensation that predicts risk of mortality in NAFLD-related cirrhosis.
© 2022 John Wiley & Sons Ltd.
Conflict of interest statement
CONFLICTS OF INTEREST
SS has no conflicts of interest. TO has no conflicts of interest. EM has no conflicts of interest. CW has no conflicts of interest. RY has no conflicts of interest. AA has no conflicts of interest. TH has no conflicts of interest. MD has no conflicts of interest. RE has no conflicts of interest. RL serves as a consultant for Anylam/Regeneron, Amgen, Arrowhead Pharmaceuticals, AstraZeneca, Bristol-Myer Squibb, CohBar, Eli Lilly, Galmed, Gilead, Glympse bio, Inipharm, Intercept, Ionis, Janssen Inc., Madrigal, Metacrine, Inc., NGM Biopharmaceuticals, Novartis, Novo Nordisk, Pfizer, Sagimet, 89 bio and Viking Therapeutics. In addition, his institution has received grant support from Allergan, Astrazeneca, Boehringer-Ingelheim, Bristol-Myers Squibb, Eli Lilly, Galectin Therapeutics, Galmed Pharmaceuticals, Genfit, Gilead, Intercept, Inventiva, Janssen, Madrigal Pharmaceuticals, Merck, NGM Biopharmaceuticals, Pfizer and Siemens. RL is also co-founder of Liponexus, Inc.
Figures

Similar articles
-
Hip Fractures in Patients With Liver Cirrhosis: Worsening Liver Function Is Associated with Increased Mortality.Clin Orthop Relat Res. 2022 Jun 1;480(6):1077-1088. doi: 10.1097/CORR.0000000000002088. Epub 2021 Dec 31. Clin Orthop Relat Res. 2022. PMID: 34978539 Free PMC article.
-
Emricasan to prevent new decompensation in patients with NASH-related decompensated cirrhosis.J Hepatol. 2021 Feb;74(2):274-282. doi: 10.1016/j.jhep.2020.09.029. Epub 2020 Oct 8. J Hepatol. 2021. PMID: 33038432 Clinical Trial.
-
Hepatic venous pressure gradient predicts clinical decompensation in patients with compensated cirrhosis.Gastroenterology. 2007 Aug;133(2):481-8. doi: 10.1053/j.gastro.2007.05.024. Epub 2007 May 21. Gastroenterology. 2007. PMID: 17681169 Clinical Trial.
-
Review article: potential mechanisms of action of rifaximin in the management of hepatic encephalopathy and other complications of cirrhosis.Aliment Pharmacol Ther. 2016 Jan;43 Suppl 1:11-26. doi: 10.1111/apt.13435. Aliment Pharmacol Ther. 2016. PMID: 26618922 Review.
-
Key Insights and Clinical Pearls in the Identification and Management of Cirrhosis and Its Complications.Am J Med. 2024 Oct;137(10):929-938. doi: 10.1016/j.amjmed.2024.05.015. Epub 2024 May 22. Am J Med. 2024. PMID: 38788826 Review.
Cited by
-
Global epidemiology of cirrhosis - aetiology, trends and predictions.Nat Rev Gastroenterol Hepatol. 2023 Jun;20(6):388-398. doi: 10.1038/s41575-023-00759-2. Epub 2023 Mar 28. Nat Rev Gastroenterol Hepatol. 2023. PMID: 36977794 Free PMC article. Review.
-
Artificial intelligence-based evaluation of prognosis in cirrhosis.J Transl Med. 2024 Oct 14;22(1):933. doi: 10.1186/s12967-024-05726-2. J Transl Med. 2024. PMID: 39402630 Free PMC article. Review.
References
-
- Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2018;15(1):11–20. - PubMed
-
- Hagström H, Nasr P, Ekstedt M, Hammar U, Stål P, Hultcrantz R, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. 2017;67(6):1265–73. - PubMed
