The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma
- PMID: 36110851
- PMCID: PMC9468372
- DOI: 10.3389/fimmu.2022.998236
The cuproptosis-related signature associated with the tumor environment and prognosis of patients with glioma
Abstract
Background: Copper ions are essential for cellular physiology. Cuproptosis is a novel method of copper-dependent cell death, and the cuproptosis-based signature for glioma remains less studied.
Methods: Several glioma datasets with clinicopathological information were collected from TCGA, GEO and CGGA. Robust Multichip Average (RMA) algorithm was used for background correction and normalization, cuproptosis-related genes (CRGs) were then collected. The TCGA-glioma cohort was clustered using ConsensusClusterPlus. Univariate Cox regression analysis and the Random Survival Forest model were performed on the differentially expressed genes to identify prognostic genes. The cuproptosis-signature was constructed by calculating CuproptosisScore using Multivariate Cox regression analysis. Differences in terms of genomic mutation, tumor microenvironment, and enrichment pathways were evaluated between high- or low-CuproptosisScore. Furthermore, drug response prediction was carried out utilizing pRRophetic.
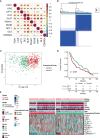
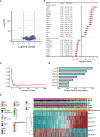
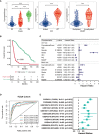
Results: Two subclusters based on CRGs were identified. Patients in cluster2 had better clinical outcomes. The cuproptosis-signature was constructed based on CuproptosisScore. Patients with higher CuproptosisScore had higher WHO grades and worse prognosis, while patients with lower grades were more likely to develop IDH mutations or MGMT methylation. Univariate and Multivariate Cox regression analysis demonstrated CuproptosisScore was an independent prognostic factor. The accuracy of the signature in prognostic prediction was further confirmed in 11 external validation datasets. In groups with high-CuproptosisScore, PIK3CA, MUC16, NF1, TTN, TP53, PTEN, and EGFR showed high mutation frequency. IDH1, TP53, ATRX, CIC, and FUBP1 demonstrated high mutation frequency in low-CuproptosisScore group. The level of immune infiltration increased as CuproptosisScore increased. SubMap analysis revealed patients with high-CuproptosisScore may respond to anti-PD-1 therapy. The IC50 values of Bexarotene, Bicalutamide, Bortezomib, and Cytarabine were lower in the high-CuproptosisScore group than those in the low-CuproptosisScore group. Finally, the importance of IGFBP2 in TCGA-glioma cohort was confirmed.
Conclusion: The current study revealed the novel cuproptosis-based signature might help predict the prognosis, biological features, and appropriate treatment for patients with glioma.
Keywords: bioinformatics; clusters; cuproptosis; glioma; signature.
Copyright © 2022 Wang, Lu, Wang, Liu, Wu, Yang, Huan and Gong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
Cuproptosis-related gene-located DNA methylation in lower-grade glioma: Prognosis and tumor microenvironment.Cancer Biomark. 2024;40(2):185-198. doi: 10.3233/CBM-230341. Cancer Biomark. 2024. PMID: 38578883 Free PMC article.
-
Signature construction and molecular subtype identification based on cuproptosis-related genes to predict the prognosis and immune activity of patients with hepatocellular carcinoma.Front Immunol. 2022 Sep 28;13:990790. doi: 10.3389/fimmu.2022.990790. eCollection 2022. Front Immunol. 2022. PMID: 36248822 Free PMC article.
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Machine learning algorithm to construct cuproptosis- and immune-related prognosis prediction model for colon cancer.World J Gastrointest Oncol. 2023 Mar 15;15(3):372-388. doi: 10.4251/wjgo.v15.i3.372. World J Gastrointest Oncol. 2023. PMID: 37009317 Free PMC article. Review.
Cited by
-
Systematic pan-cancer analysis identifies SLC31A1 as a biomarker in multiple tumor types.BMC Med Genomics. 2023 Mar 27;16(1):61. doi: 10.1186/s12920-023-01489-9. BMC Med Genomics. 2023. PMID: 36973786 Free PMC article.
-
Novel cuproptosis-related prognostic gene profiles in preeclampsia.BMC Pregnancy Childbirth. 2024 Jan 10;24(1):53. doi: 10.1186/s12884-023-06215-y. BMC Pregnancy Childbirth. 2024. PMID: 38200445 Free PMC article.
-
Identification and validation of methylation-CpG prognostic signature for prognosis of hepatocellular carcinoma.Aging (Albany NY). 2024 Jan 18;16(2):1733-1749. doi: 10.18632/aging.205454. Epub 2024 Jan 18. Aging (Albany NY). 2024. PMID: 38244582 Free PMC article.
-
Identification of molecular subtypes and a prognostic signature based on m6A/m5C/m1A-related genes in lung adenocarcinoma.Sci Rep. 2024 Mar 30;14(1):7543. doi: 10.1038/s41598-024-57910-5. Sci Rep. 2024. PMID: 38555384 Free PMC article.
-
Comprehensive bioinformatics analysis of human cytomegalovirus pathway genes in pan-cancer.Hum Genomics. 2024 Jun 17;18(1):65. doi: 10.1186/s40246-024-00633-5. Hum Genomics. 2024. PMID: 38886862 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

