Sensing of SARS-CoV-2 by pDCs and their subsequent production of IFN-I contribute to macrophage-induced cytokine storm during COVID-19
- PMID: 36083891
- PMCID: PMC9853436
- DOI: 10.1126/sciimmunol.add4906
Sensing of SARS-CoV-2 by pDCs and their subsequent production of IFN-I contribute to macrophage-induced cytokine storm during COVID-19
Abstract
Lung-infiltrating macrophages create a marked inflammatory milieu in a subset of patients with COVID-19 by producing a cytokine storm, which correlates with increased lethality. However, these macrophages are largely not infected by SARS-CoV-2, so the mechanism underlying their activation in the lung is unclear. Type I interferons (IFN-I) contribute to protecting the host against SARS-CoV-2 but may also have some deleterious effect, and the source of IFN-I in the lungs of infected patients is not well defined. Plasmacytoid dendritic cells (pDCs), a key cell type involved in antiviral responses, can produce IFN-I in response to SARS-CoV-2. We observed the infiltration of pDCs in the lungs of SARS-CoV-2-infected patients, which correlated with strong IFN-I signaling in lung macrophages. In patients with severe COVID-19, lung macrophages expressed a robust inflammatory signature, which correlated with persistent IFN-I signaling at the single-cell level. Hence, we observed the uncoupling in the kinetics of the infiltration of pDCs in the lungs and the associated IFN-I signature, with the cytokine storm in macrophages. We observed that pDCs were the dominant IFN-α-producing cells in response to the virus in the blood, whereas macrophages produced IFN-α only when in physical contact with infected epithelial cells. We also showed that IFN-α produced by pDCs, after the sensing of SARS-CoV-2 by TLR7, mediated changes in macrophages at both transcriptional and epigenetic levels, which favored their hyperactivation by environmental stimuli. Together, these data indicate that the priming of macrophages can result from the response by pDCs to SARS-CoV-2, leading to macrophage activation in patients with severe COVID-19.
Conflict of interest statement
Figures

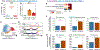
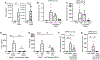

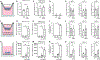

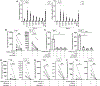

Similar articles
-
Macrophage phagocytosis of SARS-CoV-2-infected cells mediates potent plasmacytoid dendritic cell activation.Cell Mol Immunol. 2023 Jul;20(7):835-849. doi: 10.1038/s41423-023-01039-4. Epub 2023 May 30. Cell Mol Immunol. 2023. PMID: 37253946 Free PMC article.
-
Plasmacytoid dendritic cells produce type I interferon and reduce viral replication in airway epithelial cells after SARS-CoV-2 infection.bioRxiv [Preprint]. 2021 May 13:2021.05.12.443948. doi: 10.1101/2021.05.12.443948. bioRxiv. 2021. PMID: 34013278 Free PMC article. Preprint.
-
High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus.J Virol. 2015 Apr;89(7):3859-69. doi: 10.1128/JVI.03607-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609809 Free PMC article.
-
Janus kinase signaling as risk factor and therapeutic target for severe SARS-CoV-2 infection.Eur J Immunol. 2021 May;51(5):1071-1075. doi: 10.1002/eji.202149173. Epub 2021 Mar 22. Eur J Immunol. 2021. PMID: 33675065 Free PMC article. Review.
-
Plasmacytoid dendritic cells during COVID-19: Ally or adversary?Cell Rep. 2022 Jul 26;40(4):111148. doi: 10.1016/j.celrep.2022.111148. Epub 2022 Jul 14. Cell Rep. 2022. PMID: 35858624 Free PMC article. Review.
Cited by
-
Interstitial macrophages are a focus of viral takeover and inflammation in COVID-19 initiation in human lung.J Exp Med. 2024 Jun 3;221(6):e20232192. doi: 10.1084/jem.20232192. Epub 2024 Apr 10. J Exp Med. 2024. PMID: 38597954 Free PMC article.
-
The Multifaceted Functionality of Plasmacytoid Dendritic Cells in Gastrointestinal Cancers: A Potential Therapeutic Target?Cancers (Basel). 2024 Jun 13;16(12):2216. doi: 10.3390/cancers16122216. Cancers (Basel). 2024. PMID: 38927922 Free PMC article. Review.
-
Modulation of type I interferon responses potently inhibits SARS-CoV-2 replication and inflammation in rhesus macaques.Sci Immunol. 2023 Jul 28;8(85):eadg0033. doi: 10.1126/sciimmunol.adg0033. Epub 2023 Jul 28. Sci Immunol. 2023. PMID: 37506197 Free PMC article.
-
CD4+ and CD8+ T cells are required to prevent SARS-CoV-2 persistence in the nasal compartment.Sci Adv. 2024 Aug 23;10(34):eadp2636. doi: 10.1126/sciadv.adp2636. Epub 2024 Aug 23. Sci Adv. 2024. PMID: 39178263 Free PMC article.
-
Gonadal androgens are associated with decreased type I interferon production by plasmacytoid dendritic cells and increased IgG titres to BNT162b2 following co-vaccination with live attenuated influenza vaccine in adolescents.Front Immunol. 2024 Feb 28;15:1329805. doi: 10.3389/fimmu.2024.1329805. eCollection 2024. Front Immunol. 2024. PMID: 38481993 Free PMC article.
References
-
- Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, Cheng L, Li J, Wang X, Wang F, Liu L, Amit I, Zhang S, Zhang Z, Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med 26, 842–844 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

