Identification and Validation of an m6A-Related LncRNA Signature to Predict Progression-Free Survival in Colorectal Cancer
- PMID: 36032659
- PMCID: PMC9407446
- DOI: 10.3389/pore.2022.1610536
Identification and Validation of an m6A-Related LncRNA Signature to Predict Progression-Free Survival in Colorectal Cancer
Abstract
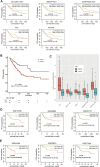
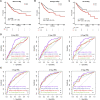
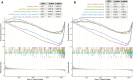
The RNA methylation of N6 adenosine (m6A) plays a crucial role in various biological processes. Strong evidence reveals that the dysregulation of long non-coding RNAs (lncRNA) brings about the abnormality of downstream signaling in multiple ways, thus influencing tumor initiation and progression. Currently, it is essential to discover effective and succinct molecular biomarkers for predicting colorectal cancer (CRC) prognosis. However, the prognostic value of m6A-related lncRNAs for CRC remains unclear, especially for progression-free survival (PFS). Here, we screened 24 m6A-related lncRNAs in 622 CRC patients and identified five lncRNAs (SLCO4A1-AS1, MELTF-AS1, SH3PXD2A-AS1, H19 and PCAT6) associated with patient PFS. Compared to normal samples, their expression was up-regulated in CRC tumors from TCGA dataset, which was validated in 55 CRC patients from our in-house cohort. We established an m6A-Lnc signature for predicting patient PFS, which was an independent prognostic factor by classification analysis of clinicopathologic features. Moreover, the signature was validated in 1,077 patients from six independent datasets (GSE17538, GSE39582, GSE33113, GSE31595, GSE29621, and GSE17536), and it showed better performance than three known lncRNA signatures for predicting PFS. In summary, our study demonstrates that the m6A-Lnc signature is a promising biomarker for forecasting patient PFS in CRC.
Keywords: colorectal cancer; lncRNA; m6A; progression free survival; signature.
Copyright © 2022 Zhang, Li, Chu, Xiao, Zhang, Li and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
N6-Methyladenosine-Related lncRNA Signature Predicts the Overall Survival of Colorectal Cancer Patients.Genes (Basel). 2021 Aug 31;12(9):1375. doi: 10.3390/genes12091375. Genes (Basel). 2021. PMID: 34573357 Free PMC article.
-
Comprehensive Analysis of N6-Methyladenosine-Related lncRNA Signature for Predicting Prognosis and Immune Cell Infiltration in Patients with Colorectal Cancer.Dis Markers. 2021 Oct 28;2021:8686307. doi: 10.1155/2021/8686307. eCollection 2021. Dis Markers. 2021. PMID: 34745388 Free PMC article.
-
Discovery of a novel six-long non-coding RNA signature predicting survival of colorectal cancer patients.J Cell Biochem. 2018 Apr;119(4):3574-3585. doi: 10.1002/jcb.26548. Epub 2018 Jan 15. J Cell Biochem. 2018. PMID: 29227531
-
Identification of N6-methylandenosine related lncRNA signatures for predicting the prognosis and therapy response in colorectal cancer patients.Front Genet. 2022 Sep 30;13:947747. doi: 10.3389/fgene.2022.947747. eCollection 2022. Front Genet. 2022. PMID: 36246627 Free PMC article.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
Cited by
-
Prognostic significance of a signature based on senescence-related genes in colorectal cancer.Geroscience. 2024 Oct;46(5):4495-4504. doi: 10.1007/s11357-024-01164-6. Epub 2024 Apr 25. Geroscience. 2024. PMID: 38658505 Free PMC article.
-
The lncRNA epigenetics: The significance of m6A and m5C lncRNA modifications in cancer.Front Oncol. 2023 Mar 9;13:1063636. doi: 10.3389/fonc.2023.1063636. eCollection 2023. Front Oncol. 2023. PMID: 36969033 Free PMC article. Review.
References
-
- Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-Years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol (2017) 3(4):524–48. 10.1001/jamaoncol.2016.5688 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

