Increasing cell size remodels the proteome and promotes senescence
- PMID: 35987199
- PMCID: PMC9444988
- DOI: 10.1016/j.molcel.2022.07.017
Increasing cell size remodels the proteome and promotes senescence
Abstract
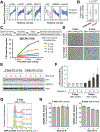
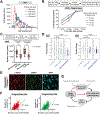
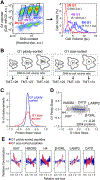
Cell size is tightly controlled in healthy tissues, but it is unclear how deviations in cell size affect cell physiology. To address this, we measured how the cell's proteome changes with increasing cell size. Size-dependent protein concentration changes are widespread and predicted by subcellular localization, size-dependent mRNA concentrations, and protein turnover. As proliferating cells grow larger, concentration changes typically associated with cellular senescence are increasingly pronounced, suggesting that large size may be a cause rather than just a consequence of cell senescence. Consistent with this hypothesis, larger cells are prone to replicative, DNA-damage-induced, and CDK4/6i-induced senescence. Size-dependent changes to the proteome, including those associated with senescence, are not observed when an increase in cell size is accompanied by an increase in ploidy. Together, our findings show how cell size could impact many aspects of cell physiology by remodeling the proteome and provide a rationale for cell size control and polyploidization.
Keywords: DNA damage; SA-beta-Gal; cell cycle; cell size; p16(INK4); palbociclib; polyploidy; proteomics; senescence; size-scaling.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
Delineation of proteome changes driven by cell size and growth rate.Front Cell Dev Biol. 2022 Sep 5;10:980721. doi: 10.3389/fcell.2022.980721. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36133920 Free PMC article.
-
Expression of senescence-associated beta-galactosidase in enlarged prostates from men with benign prostatic hyperplasia.Urology. 2000 Jul;56(1):160-6. doi: 10.1016/s0090-4295(00)00538-0. Urology. 2000. PMID: 10869659
-
Techniques to Induce and Quantify Cellular Senescence.J Vis Exp. 2017 May 1;(123):55533. doi: 10.3791/55533. J Vis Exp. 2017. PMID: 28518126 Free PMC article.
-
What is and what is not cell senescence.Postepy Biochem. 2018 Oct 15;64(2):110-118. doi: 10.18388/pb.2018_120. Postepy Biochem. 2018. PMID: 30656893 Review. English.
-
The role of autophagy in escaping therapy-induced polyploidy/senescence.Adv Cancer Res. 2021;150:209-247. doi: 10.1016/bs.acr.2021.01.004. Epub 2021 Mar 11. Adv Cancer Res. 2021. PMID: 33858597 Review.
Cited by
-
The cell cycle and cell size influence the rates of global cellular translation and transcription in fission yeast.EMBO J. 2023 May 2;42(9):e113333. doi: 10.15252/embj.2022113333. Epub 2023 Mar 23. EMBO J. 2023. PMID: 36951016 Free PMC article.
-
Yearning for machine learning: applications for the classification and characterisation of senescence.Cell Tissue Res. 2023 Oct;394(1):1-16. doi: 10.1007/s00441-023-03768-4. Epub 2023 Apr 5. Cell Tissue Res. 2023. PMID: 37016180 Free PMC article. Review.
-
Delineation of proteome changes driven by cell size and growth rate.Front Cell Dev Biol. 2022 Sep 5;10:980721. doi: 10.3389/fcell.2022.980721. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36133920 Free PMC article.
-
Repeated Administration of Cisplatin Transforms Kidney Fibroblasts through G2/M Arrest and Cellular Senescence.Cells. 2022 Nov 2;11(21):3472. doi: 10.3390/cells11213472. Cells. 2022. PMID: 36359868 Free PMC article.
-
The Nuclear-to-Cytoplasmic Ratio: Coupling DNA Content to Cell Size, Cell Cycle, and Biosynthetic Capacity.Annu Rev Genet. 2022 Nov 30;56:165-185. doi: 10.1146/annurev-genet-080320-030537. Epub 2022 Aug 17. Annu Rev Genet. 2022. PMID: 35977407 Free PMC article. Review.
References
-
- Berry S, Muller M, Rai A, and Pelkmans L (2022). Feedback from nuclear RNA on transcription promotes robust RNA concentration homeostasis in human cells. Cell Syst. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

