Islet Gene View-a tool to facilitate islet research
- PMID: 35948367
- PMCID: PMC9366203
- DOI: 10.26508/lsa.202201376
Islet Gene View-a tool to facilitate islet research
Abstract
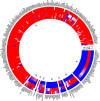
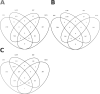
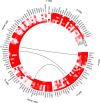
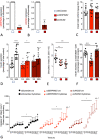
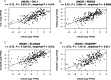
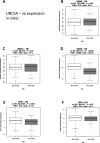
Characterization of gene expression in pancreatic islets and its alteration in type 2 diabetes (T2D) are vital in understanding islet function and T2D pathogenesis. We leveraged RNA sequencing and genome-wide genotyping in islets from 188 donors to create the Islet Gene View (IGW) platform to make this information easily accessible to the scientific community. Expression data were related to islet phenotypes, diabetes status, other islet-expressed genes, islet hormone-encoding genes and for expression in insulin target tissues. The IGW web application produces output graphs for a particular gene of interest. In IGW, 284 differentially expressed genes (DEGs) were identified in T2D donor islets compared with controls. Forty percent of DEGs showed cell-type enrichment and a large proportion significantly co-expressed with islet hormone-encoding genes; glucagon (<i>GCG</i>, 56%), amylin (<i>IAPP</i>, 52%), insulin (<i>INS</i>, 44%), and somatostatin (<i>SST</i>, 24%). Inhibition of two DEGs, <i>UNC5D</i> and <i>SERPINE2</i>, impaired glucose-stimulated insulin secretion and impacted cell survival in a human β-cell model. The exploratory use of IGW could help designing more comprehensive functional follow-up studies and serve to identify therapeutic targets in T2D.
© 2022 Asplund et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures










Similar articles
-
Altered serotonin (5-HT) 1D and 2A receptor expression may contribute to defective insulin and glucagon secretion in human type 2 diabetes.Peptides. 2015 Sep;71:113-20. doi: 10.1016/j.peptides.2015.07.008. Epub 2015 Jul 20. Peptides. 2015. PMID: 26206285
-
Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis.Diabetes. 2017 Apr;66(4):1074-1085. doi: 10.2337/db16-0996. Epub 2017 Jan 4. Diabetes. 2017. PMID: 28052964
-
Nesfatin-1 stimulates glucagon and insulin secretion and beta cell NUCB2 is reduced in human type 2 diabetic subjects.Cell Tissue Res. 2011 Dec;346(3):393-405. doi: 10.1007/s00441-011-1268-5. Epub 2011 Nov 23. Cell Tissue Res. 2011. PMID: 22108805
-
Islet amyloid polypeptide in the islets of Langerhans: friend or foe?Diabetologia. 2000 Jun;43(6):687-95. doi: 10.1007/s001250051364. Diabetologia. 2000. PMID: 10907112 Review.
-
Modulation of the pancreatic islet-stress axis as a novel potential therapeutic target in diabetes mellitus.Vitam Horm. 2014;95:195-222. doi: 10.1016/B978-0-12-800174-5.00008-9. Vitam Horm. 2014. PMID: 24559919 Review.
Cited by
-
The highly expressed calcium-insensitive synaptotagmin-11 and synaptotagmin-13 modulate insulin secretion.Acta Physiol (Oxf). 2022 Sep;236(1):e13857. doi: 10.1111/apha.13857. Epub 2022 Jul 2. Acta Physiol (Oxf). 2022. PMID: 35753051 Free PMC article.
-
The Calcium Channel Subunit Gamma-4 as a Novel Regulator of MafA in Pancreatic Beta-Cell Controls Glucose Homeostasis.Biomedicines. 2022 Mar 25;10(4):770. doi: 10.3390/biomedicines10040770. Biomedicines. 2022. PMID: 35453520 Free PMC article.
-
Ribosomal biogenesis regulator DIMT1 controls β-cell protein synthesis, mitochondrial function, and insulin secretion.J Biol Chem. 2022 Mar;298(3):101692. doi: 10.1016/j.jbc.2022.101692. Epub 2022 Feb 8. J Biol Chem. 2022. PMID: 35148993 Free PMC article.
-
Intracellular C3 protects β-cells from IL-1β-driven cytotoxicity via interaction with Fyn-related kinase.Proc Natl Acad Sci U S A. 2024 Feb 20;121(8):e2312621121. doi: 10.1073/pnas.2312621121. Epub 2024 Feb 12. Proc Natl Acad Sci U S A. 2024. PMID: 38346191 Free PMC article.
-
Investigating the Impact of IL6 on Insulin Secretion: Evidence from INS-1 Cells, Human Pancreatic Islets, and Serum Analysis.Cells. 2024 Apr 15;13(8):685. doi: 10.3390/cells13080685. Cells. 2024. PMID: 38667300 Free PMC article.
References
-
- Ahlqvist E, Storm P, Karajamaki A, Martinell M, Dorkhan M, Carlsson A, Vikman P, Prasad RB, Aly DM, Almgren P, et al. (2018) Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6: 361–369. 10.1016/S2213-8587(18)30051-2 - DOI - PubMed
-
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300. 10.1111/J.2517-6161.1995.TB02031.X - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
