Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface
- PMID: 35913979
- PMCID: PMC9371267
- DOI: 10.1371/journal.pbio.3001714
Structure of the human galanin receptor 2 bound to galanin and Gq reveals the basis of ligand specificity and how binding affects the G-protein interface
Abstract
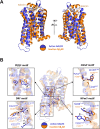
Galanin is a neuropeptide expressed in the central and peripheral nervous systems, where it regulates various processes including neuroendocrine release, cognition, and nerve regeneration. Three G-protein coupled receptors (GPCRs) for galanin have been discovered, which is the focus of efforts to treat diseases including Alzheimer's disease, anxiety, and addiction. To understand the basis of the ligand preferences of the receptors and to assist structure-based drug design, we used cryo-electron microscopy (cryo-EM) to solve the molecular structure of GALR2 bound to galanin and a cognate heterotrimeric G-protein, providing a molecular view of the neuropeptide binding site. Mutant proteins were assayed to help reveal the basis of ligand specificity, and structural comparison between the activated GALR2 and inactive hβ2AR was used to relate galanin binding to the movements of transmembrane (TM) helices and the G-protein interface.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Theoretical investigation of selective ligand binding mode of galanin receptors.J Biomol Struct Dyn. 2022;40(23):12964-12974. doi: 10.1080/07391102.2021.1977703. Epub 2021 Oct 11. J Biomol Struct Dyn. 2022. PMID: 34632940
-
Homology modeling, docking, and molecular dynamics simulation of the receptor GALR2 and its interactions with galanin and a positive allosteric modulator.J Mol Model. 2016 Apr;22(4):90. doi: 10.1007/s00894-016-2944-x. Epub 2016 Mar 28. J Mol Model. 2016. PMID: 27021209
-
Coevolution of the spexin/galanin/kisspeptin family: Spexin activates galanin receptor type II and III.Endocrinology. 2014 May;155(5):1864-73. doi: 10.1210/en.2013-2106. Epub 2014 Feb 11. Endocrinology. 2014. PMID: 24517231
-
Galanin, galanin receptors, and drug targets.Exp Suppl. 2010;102:7-23. doi: 10.1007/978-3-0346-0228-0_2. Exp Suppl. 2010. PMID: 21299058 Review.
-
Galanin, galanin receptors and drug targets.Cell Mol Life Sci. 2008 Jun;65(12):1796-805. doi: 10.1007/s00018-008-8153-8. Cell Mol Life Sci. 2008. PMID: 18500647 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

