Increasing the resilience of plant immunity to a warming climate
- PMID: 35768511
- PMCID: PMC9279160
- DOI: 10.1038/s41586-022-04902-y
Increasing the resilience of plant immunity to a warming climate
Abstract
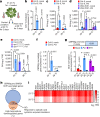
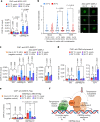
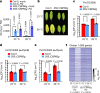
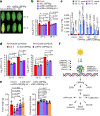
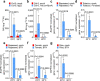
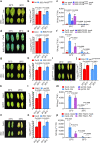
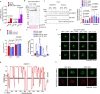
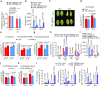
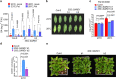
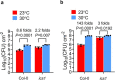
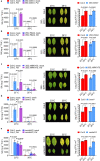
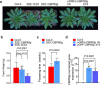
Extreme weather conditions associated with climate change affect many aspects of plant and animal life, including the response to infectious diseases. Production of salicylic acid (SA), a central plant defence hormone1-3, is particularly vulnerable to suppression by short periods of hot weather above the normal plant growth temperature range via an unknown mechanism4-7. Here we show that suppression of SA production in Arabidopsis thaliana at 28 °C is independent of PHYTOCHROME B8,9 (phyB) and EARLY FLOWERING 310 (ELF3), which regulate thermo-responsive plant growth and development. Instead, we found that formation of GUANYLATE BINDING PROTEIN-LIKE 3 (GBPL3) defence-activated biomolecular condensates11 (GDACs) was reduced at the higher growth temperature. The altered GDAC formation in vivo is linked to impaired recruitment of GBPL3 and SA-associated Mediator subunits to the promoters of CBP60g and SARD1, which encode master immune transcription factors. Unlike many other SA signalling components, including the SA receptor and biosynthetic genes, optimized CBP60g expression was sufficient to broadly restore SA production, basal immunity and effector-triggered immunity at the elevated growth temperature without significant growth trade-offs. CBP60g family transcription factors are widely conserved in plants12. These results have implications for safeguarding the plant immune system as well as understanding the concept of the plant-pathogen-environment disease triangle and the emergence of new disease epidemics in a warming climate.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Comment in
-
Shedding light on immune suppression at high temperature.Trends Microbiol. 2022 Oct;30(10):918-919. doi: 10.1016/j.tim.2022.07.010. Epub 2022 Aug 3. Trends Microbiol. 2022. PMID: 35931623
-
Sustaining plant immunity in rising temperature.Cell Res. 2022 Dec;32(12):1038-1039. doi: 10.1038/s41422-022-00710-1. Cell Res. 2022. PMID: 35931822 Free PMC article. No abstract available.
-
Too hot to defend: a tale of salicylic acid.Trends Plant Sci. 2023 Jan;28(1):4-6. doi: 10.1016/j.tplants.2022.10.001. Epub 2022 Oct 19. Trends Plant Sci. 2023. PMID: 36272889
Similar articles
-
Distinct profiles of plant immune resilience revealed by natural variation in warm temperature-modulated disease resistance among Arabidopsis accessions.Plant Cell Environ. 2024 Dec;47(12):5115-5125. doi: 10.1111/pce.15098. Epub 2024 Aug 20. Plant Cell Environ. 2024. PMID: 39165012
-
Structural diversity and stress regulation of the plant immunity-associated CALMODULIN-BINDING PROTEIN 60 (CBP60) family of transcription factors in Solanum lycopersicum (tomato).Funct Integr Genomics. 2023 Jul 13;23(3):236. doi: 10.1007/s10142-023-01172-3. Funct Integr Genomics. 2023. PMID: 37439880
-
TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g).New Phytol. 2018 Jan;217(1):344-354. doi: 10.1111/nph.14780. Epub 2017 Sep 12. New Phytol. 2018. PMID: 28898429
-
Molecular regulation of the salicylic acid hormone pathway in plants under changing environmental conditions.Trends Biochem Sci. 2023 Aug;48(8):699-712. doi: 10.1016/j.tibs.2023.05.004. Epub 2023 May 29. Trends Biochem Sci. 2023. PMID: 37258325 Review.
-
NPR1 in JazzSet with Pathogen Effectors.Trends Plant Sci. 2018 Jun;23(6):469-472. doi: 10.1016/j.tplants.2018.04.007. Epub 2018 May 9. Trends Plant Sci. 2018. PMID: 29753632 Review.
Cited by
-
Response of root and root hair phenotypes of cotton seedlings under high temperature revealed with RhizoPot.Front Plant Sci. 2022 Nov 8;13:1007145. doi: 10.3389/fpls.2022.1007145. eCollection 2022. Front Plant Sci. 2022. PMID: 36426149 Free PMC article.
-
Stress-related biomolecular condensates in plants.Plant Cell. 2023 Sep 1;35(9):3187-3204. doi: 10.1093/plcell/koad127. Plant Cell. 2023. PMID: 37162152 Free PMC article.
-
Climate change impacts on plant pathogens, food security and paths forward.Nat Rev Microbiol. 2023 Oct;21(10):640-656. doi: 10.1038/s41579-023-00900-7. Epub 2023 May 2. Nat Rev Microbiol. 2023. PMID: 37131070 Free PMC article. Review.
-
Is there a specific calcium signal out there to decode combined biotic stress and temperature elevation?Front Plant Sci. 2022 Nov 3;13:1004406. doi: 10.3389/fpls.2022.1004406. eCollection 2022. Front Plant Sci. 2022. PMID: 36407594 Free PMC article. No abstract available.
-
Liquid-liquid phase separation in plants: Advances and perspectives from model species to crops.Plant Commun. 2024 Jan 8;5(1):100663. doi: 10.1016/j.xplc.2023.100663. Epub 2023 Jul 26. Plant Commun. 2024. PMID: 37496271 Free PMC article. Review.
References
-
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. - PubMed
-
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 2013;64:839–863. - PubMed
-
- Peng Y, Yang J, Li X, Zhang Y. Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 2021;72:761–791. - PubMed
-
- Castroverde CDM, Dina D. Temperature regulation of plant hormone signaling during stress and development. J. Exp. Bot. 2021;72:7436–7458. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

