Extended intergenic DNA contributes to neuron-specific expression of neighboring genes in the mammalian nervous system
- PMID: 35585070
- PMCID: PMC9117226
- DOI: 10.1038/s41467-022-30192-z
Extended intergenic DNA contributes to neuron-specific expression of neighboring genes in the mammalian nervous system
Abstract
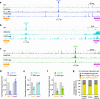
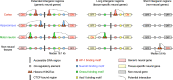
Mammalian genomes comprise largely intergenic noncoding DNA with numerous cis-regulatory elements. Whether and how the size of intergenic DNA affects gene expression in a tissue-specific manner remain unknown. Here we show that genes with extended intergenic regions are preferentially expressed in neural tissues but repressed in other tissues in mice and humans. Extended intergenic regions contain twice as many active enhancers in neural tissues compared to other tissues. Neural genes with extended intergenic regions are globally co-expressed with neighboring neural genes controlled by distinct enhancers in the shared intergenic regions. Moreover, generic neural genes expressed in multiple tissues have significantly longer intergenic regions than neural genes expressed in fewer tissues. The intergenic regions of the generic neural genes have many tissue-specific active enhancers containing distinct transcription factor binding sites specific to each neural tissue. We also show that genes with extended intergenic regions are enriched for neural genes only in vertebrates. The expansion of intergenic regions may reflect the regulatory complexity of tissue-type-specific gene expression in the nervous system.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Unravelling cis-regulatory elements in the genome of the smallest photosynthetic eukaryote: phylogenetic footprinting in Ostreococcus.J Mol Evol. 2009 Sep;69(3):249-59. doi: 10.1007/s00239-009-9271-0. Epub 2009 Aug 20. J Mol Evol. 2009. PMID: 19693423
-
Decoding cis-regulatory systems in ascidians.Zoolog Sci. 2005 Feb;22(2):129-46. doi: 10.2108/zsj.22.129. Zoolog Sci. 2005. PMID: 15738634 Review.
-
Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites.Dev Biol. 2011 Sep 15;357(2):450-62. doi: 10.1016/j.ydbio.2011.03.007. Epub 2011 Mar 22. Dev Biol. 2011. PMID: 21435340 Free PMC article.
-
Functions of noncoding sequences in mammalian genomes.Biochemistry (Mosc). 2014 Dec;79(13):1442-69. doi: 10.1134/S0006297914130021. Biochemistry (Mosc). 2014. PMID: 25749159 Review.
-
Sequence analysis of a Hoxa4-Hoxa5 intergenic region including shared regulatory elements.DNA Seq. 2002 Aug;13(4):203-9. doi: 10.1080/10425170290034507. DNA Seq. 2002. PMID: 12487022
Cited by
-
Improving rigor and reproducibility in chromatin immunoprecipitation assay data analysis workflows with Rocketchip.bioRxiv [Preprint]. 2024 Jul 16:2024.07.10.602975. doi: 10.1101/2024.07.10.602975. bioRxiv. 2024. PMID: 39071274 Free PMC article. Preprint.
-
Genome-wide Methylation Dynamics and Context-dependent Gene Expression Variability in Differentiating Preadipocytes.J Endocr Soc. 2024 Jun 27;8(8):bvae121. doi: 10.1210/jendso/bvae121. eCollection 2024 Jul 1. J Endocr Soc. 2024. PMID: 38966711 Free PMC article.
-
From compartments to loops: understanding the unique chromatin organization in neuronal cells.Epigenetics Chromatin. 2024 May 23;17(1):18. doi: 10.1186/s13072-024-00538-6. Epigenetics Chromatin. 2024. PMID: 38783373 Free PMC article. Review.
-
Whole genome methylation sequencing reveals epigenetic landscape and abnormal expression of FABP5 in extramammary Paget's disease.Skin Res Technol. 2023 Oct;29(10):e13497. doi: 10.1111/srt.13497. Skin Res Technol. 2023. PMID: 37881057 Free PMC article.
-
A genome-wide association study (GWAS) of the personality constructs in CPAI-2 in Taiwanese Hakka populations.PLoS One. 2023 Feb 17;18(2):e0281903. doi: 10.1371/journal.pone.0281903. eCollection 2023. PLoS One. 2023. PMID: 36800362 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

