Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination
- PMID: 35584773
- PMCID: PMC9279157
- DOI: 10.1038/s41586-022-04865-0
Limited cross-variant immunity from SARS-CoV-2 Omicron without vaccination
Abstract
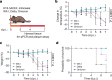
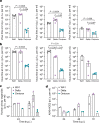
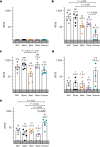
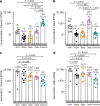
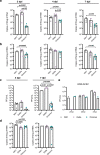
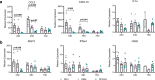
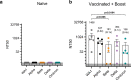
SARS-CoV-2 Delta and Omicron are globally relevant variants of concern. Although individuals infected with Delta are at risk of developing severe lung disease, infection with Omicron often causes milder symptoms, especially in vaccinated individuals1,2. The question arises of whether widespread Omicron infections could lead to future cross-variant protection, accelerating the end of the pandemic. Here we show that without vaccination, infection with Omicron induces a limited humoral immune response in mice and humans. Sera from mice overexpressing the human ACE2 receptor and infected with Omicron neutralize only Omicron, but not other variants of concern, whereas broader cross-variant neutralization was observed after WA1 and Delta infections. Unlike WA1 and Delta, Omicron replicates to low levels in the lungs and brains of infected animals, leading to mild disease with reduced expression of pro-inflammatory cytokines and diminished activation of lung-resident T cells. Sera from individuals who were unvaccinated and infected with Omicron show the same limited neutralization of only Omicron itself. By contrast, Omicron breakthrough infections induce overall higher neutralization titres against all variants of concern. Our results demonstrate that Omicron infection enhances pre-existing immunity elicited by vaccines but, on its own, may not confer broad protection against non-Omicron variants in unvaccinated individuals.
© 2022. The Author(s).
Conflict of interest statement
J.A.D. is a cofounder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics, Intellia Therapeutics and Mammoth Biosciences; a scientific advisory board member of Vertex, Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, Algen Biotechnologies, Felix Biosciences, The Column Group and Inari; a director at Johnson & Johnson and Tempus; and has research projects sponsored by Biogen, Pfizer, AppleTree Partners and Roche. C.Y.C. is the director of the UCSF-Abbott Viral Diagnostics and Discovery Study; receives research support from Abbott Laboratories; and also receives support for SARS-CoV-2 research unrelated to this study from Mammoth Biosciences.
Figures






Update of
-
Limited Cross-Variant Immunity after Infection with the SARS-CoV-2 Omicron Variant Without Vaccination.medRxiv [Preprint]. 2022 Feb 9:2022.01.13.22269243. doi: 10.1101/2022.01.13.22269243. medRxiv. 2022. Update in: Nature. 2022 Jul;607(7918):351-355. doi: 10.1038/s41586-022-04865-0 PMID: 35075459 Free PMC article. Updated. Preprint.
Comment in
-
Limited cross-variant neutralization after primary Omicron infection: consideration for a variant-containing booster.Signal Transduct Target Ther. 2022 Aug 22;7(1):294. doi: 10.1038/s41392-022-01146-0. Signal Transduct Target Ther. 2022. PMID: 35995763 Free PMC article. No abstract available.
Similar articles
-
Limited Cross-Variant Immunity after Infection with the SARS-CoV-2 Omicron Variant Without Vaccination.medRxiv [Preprint]. 2022 Feb 9:2022.01.13.22269243. doi: 10.1101/2022.01.13.22269243. medRxiv. 2022. Update in: Nature. 2022 Jul;607(7918):351-355. doi: 10.1038/s41586-022-04865-0 PMID: 35075459 Free PMC article. Updated. Preprint.
-
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone.Int J Mol Sci. 2022 Jul 12;23(14):7675. doi: 10.3390/ijms23147675. Int J Mol Sci. 2022. PMID: 35887023 Free PMC article.
-
Delta Infection After Vaccination Elicits Potent Neutralizing Immunity Against Severe Acute Respiratory Syndrome Coronavirus 2 Omicron.J Infect Dis. 2022 Nov 1;226(9):1551-1555. doi: 10.1093/infdis/jiac149. J Infect Dis. 2022. PMID: 35429398 Free PMC article.
-
Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost.Emerg Microbes Infect. 2022 Dec;11(1):337-343. doi: 10.1080/22221751.2021.2022440. Emerg Microbes Infect. 2022. PMID: 34935594 Free PMC article.
-
Spotlights on the latest opinions on identification, prevention, and management of newer CoV-2 variants: A roundup appraisal on innovative ideas and designer vaccines for Omicron.Transfus Apher Sci. 2022 Dec;61(6):103499. doi: 10.1016/j.transci.2022.103499. Epub 2022 Jul 3. Transfus Apher Sci. 2022. PMID: 35811273 Free PMC article. Review.
Cited by
-
Role of SARS-CoV-2-induced cytokine storm in multi-organ failure: Molecular pathways and potential therapeutic options.Int Immunopharmacol. 2022 Dec;113(Pt B):109428. doi: 10.1016/j.intimp.2022.109428. Epub 2022 Nov 7. Int Immunopharmacol. 2022. PMID: 36379152 Free PMC article. Review.
-
A self-assembled trimeric protein vaccine induces protective immunity against Omicron variant.Nat Commun. 2022 Sep 17;13(1):5459. doi: 10.1038/s41467-022-33209-9. Nat Commun. 2022. PMID: 36115859 Free PMC article.
-
Would New SARS-CoV-2 Variants Change the War against COVID-19?Epidemiologia (Basel). 2022 Apr 29;3(2):229-237. doi: 10.3390/epidemiologia3020018. Epidemiologia (Basel). 2022. PMID: 36417254 Free PMC article.
-
S Trimer Derived from SARS-CoV-2 B.1.351 and B.1.618 Induced Effective Immune Response against Multiple SARS-CoV-2 Variants.Vaccines (Basel). 2023 Jan 16;11(1):193. doi: 10.3390/vaccines11010193. Vaccines (Basel). 2023. PMID: 36680037 Free PMC article.
-
A fourth dose of Omicron RBD vaccine enhances broad neutralization against SARS-CoV-2 variants including BA.1 and BA.2 in vaccinated mice.J Med Virol. 2022 Aug;94(8):3992-3997. doi: 10.1002/jmv.27811. Epub 2022 May 6. J Med Virol. 2022. PMID: 35474319 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

