One AP2/ERF Transcription Factor Positively Regulates Pi Uptake and Drought Tolerance in Poplar
- PMID: 35563632
- PMCID: PMC9099566
- DOI: 10.3390/ijms23095241
One AP2/ERF Transcription Factor Positively Regulates Pi Uptake and Drought Tolerance in Poplar
Abstract
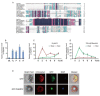
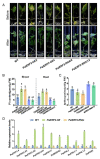
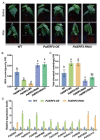
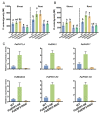
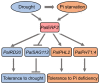
Drought decreases the inorganic phosphate (Pi) supply of soil, resulting in Pi starvation of plants, but the molecular mechanism of how plants, especially the perennial trees, are tolerant to drought stress and Pi starvation, is still elusive. In this study, we identified an AP2/ERF transcription factor gene, PalERF2, from Populus alba var. pyramidalis, and it was induced by both mannitol treatment and Pi starvation. Overexpressing and knocking-down of PalERF2 both enhanced and attenuated tolerance to drought stress and Pi deficiency compared to WT, respectively. Moreover, the overexpression of PalERF2 up-regulated the expression levels of Pi starvation-induced (PSI) genes and increased Pi uptake under drought conditions; however, its RNAi poplar showed the opposite phenotypes. Subsequent analysis indicated that PalERF2 directly modulated expressions of drought-responsive genes PalRD20 and PalSAG113, as well as PSI genes PalPHL2 and PalPHT1;4, through binding to the DRE motifs on their promoters. These results clearly indicate that poplars can recruit PalERF2 to increase the tolerance to drought and also elevate Pi uptake under drought stress.
Keywords: PalERF2; Populus; drought stress; inorganic phosphate starvation; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
PtaERF194 inhibits plant growth and enhances drought tolerance in poplar.Tree Physiol. 2022 Aug 6;42(8):1678-1692. doi: 10.1093/treephys/tpac026. Tree Physiol. 2022. PMID: 35220440
-
Functional analyses of NDPK2 in Populus trichocarpa and overexpression of PtNDPK2 enhances growth and tolerance to abiotic stresses in transgenic poplar.Plant Physiol Biochem. 2017 Aug;117:61-74. doi: 10.1016/j.plaphy.2017.05.019. Epub 2017 May 31. Plant Physiol Biochem. 2017. PMID: 28587994
-
Transcription Factor ERF194 Modulates the Stress-Related Physiology to Enhance Drought Tolerance of Poplar.Int J Mol Sci. 2023 Jan 2;24(1):788. doi: 10.3390/ijms24010788. Int J Mol Sci. 2023. PMID: 36614232 Free PMC article.
-
The C2 H2 -type zinc finger transcription factor OSIC1 positively regulates stomatal closure under osmotic stress in poplar.Plant Biotechnol J. 2023 May;21(5):943-960. doi: 10.1111/pbi.14007. Epub 2023 Feb 4. Plant Biotechnol J. 2023. PMID: 36632734 Free PMC article.
-
Cross-talk between Phosphate Starvation and Other Environmental Stress Signaling Pathways in Plants.Mol Cells. 2017 Oct;40(10):697-705. doi: 10.14348/molcells.2017.0192. Epub 2017 Oct 17. Mol Cells. 2017. PMID: 29047263 Free PMC article. Review.
Cited by
-
ZmARF1 positively regulates low phosphorus stress tolerance via modulating lateral root development in maize.PLoS Genet. 2024 Feb 5;20(2):e1011135. doi: 10.1371/journal.pgen.1011135. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38315718 Free PMC article.
-
Characterization of bZIP Transcription Factors in Transcriptome of Chrysanthemum mongolicum and Roles of CmbZIP9 in Drought Stress Resistance.Plants (Basel). 2024 Jul 26;13(15):2064. doi: 10.3390/plants13152064. Plants (Basel). 2024. PMID: 39124182 Free PMC article.
-
Transcriptomic and biochemical analyses of drought response mechanism in mung bean (Vignaradiata (L.) Wilczek) leaves.PLoS One. 2023 May 10;18(5):e0285400. doi: 10.1371/journal.pone.0285400. eCollection 2023. PLoS One. 2023. PMID: 37163521 Free PMC article.
-
ERF subfamily transcription factors and their function in plant responses to abiotic stresses.Front Plant Sci. 2022 Nov 30;13:1042084. doi: 10.3389/fpls.2022.1042084. eCollection 2022. Front Plant Sci. 2022. PMID: 36531407 Free PMC article. Review.
-
Understanding AP2/ERF Transcription Factor Responses and Tolerance to Various Abiotic Stresses in Plants: A Comprehensive Review.Int J Mol Sci. 2024 Jan 11;25(2):893. doi: 10.3390/ijms25020893. Int J Mol Sci. 2024. PMID: 38255967 Free PMC article. Review.
References
-
- López-Bucio J., Hernández-Abreu E., Sánchez-Calderón L., Nieto-Jacobo M.F., Simpson J., Herrera-Estrella L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002;129:244–256. doi: 10.1104/pp.010934. - DOI - PMC - PubMed
-
- Staaf H. Plant nutrient changes in beech leaves during senescence as influenced by site characteristics. Acta. Oecol. Oec. Plant. 1982;3:161–170.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

