A targetable CoQ-FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers
- PMID: 35459868
- PMCID: PMC9033817
- DOI: 10.1038/s41467-022-29905-1
A targetable CoQ-FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers
Abstract
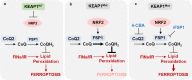
Targeting ferroptosis, a unique cell death modality triggered by unrestricted lipid peroxidation, in cancer therapy is hindered by our incomplete understanding of ferroptosis mechanisms under specific cancer genetic contexts. KEAP1 (kelch-like ECH associated protein 1) is frequently mutated or inactivated in lung cancers, and KEAP1 mutant lung cancers are refractory to most therapies, including radiotherapy. In this study, we identify ferroptosis suppressor protein 1 (FSP1, also known as AIFM2) as a transcriptional target of nuclear factor erythroid 2-related factor 2 (NRF2) and reveal that the ubiquinone (CoQ)-FSP1 axis mediates ferroptosis- and radiation- resistance in KEAP1 deficient lung cancer cells. We further show that pharmacological inhibition of the CoQ-FSP1 axis sensitizes KEAP1 deficient lung cancer cells or patient-derived xenograft tumors to radiation through inducing ferroptosis. Together, our study identifies CoQ-FSP1 as a key downstream effector of KEAP1-NRF2 pathway and as a potential therapeutic target for treating KEAP1 mutant lung cancers.
© 2022. The Author(s).
Conflict of interest statement
K.O. is a full-time employee of the Barer Institute and a former full-time employee of Kadmon Corporation. L.K. and M.V.P. are full-time employees of Kadmon Corporation.
Figures





Similar articles
-
FSP1, a novel KEAP1/NRF2 target gene regulating ferroptosis and radioresistance in lung cancers.Oncotarget. 2022 Oct 19;13:1136-1139. doi: 10.18632/oncotarget.28301. Oncotarget. 2022. PMID: 36264074 Free PMC article.
-
FSP1 confers ferroptosis resistance in KEAP1 mutant non-small cell lung carcinoma in NRF2-dependent and -independent manner.Cell Death Dis. 2023 Aug 26;14(8):567. doi: 10.1038/s41419-023-06070-x. Cell Death Dis. 2023. PMID: 37633973 Free PMC article.
-
The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis.Nature. 2019 Nov;575(7784):688-692. doi: 10.1038/s41586-019-1705-2. Epub 2019 Oct 21. Nature. 2019. PMID: 31634900 Free PMC article.
-
Targeting Nrf2 for ferroptosis-based therapy: Implications for overcoming ferroptosis evasion and therapy resistance in cancer.Biochim Biophys Acta Mol Basis Dis. 2023 Oct;1869(7):166788. doi: 10.1016/j.bbadis.2023.166788. Epub 2023 Jun 10. Biochim Biophys Acta Mol Basis Dis. 2023. PMID: 37302427 Review.
-
The crosstalk between classic cell signaling pathways, non-coding RNAs and ferroptosis in drug resistance of tumors.Cell Signal. 2023 Feb;102:110538. doi: 10.1016/j.cellsig.2022.110538. Epub 2022 Nov 24. Cell Signal. 2023. PMID: 36436800 Review.
Cited by
-
FGF21 inhibits ferroptosis caused by mitochondrial damage to promote the repair of peripheral nerve injury.Front Pharmacol. 2024 Sep 23;15:1358646. doi: 10.3389/fphar.2024.1358646. eCollection 2024. Front Pharmacol. 2024. PMID: 39376607 Free PMC article.
-
Ferroptosis in Ischemic Stroke and Related Traditional Chinese Medicines.Molecules. 2024 Sep 13;29(18):4359. doi: 10.3390/molecules29184359. Molecules. 2024. PMID: 39339354 Free PMC article. Review.
-
The cell biology of ferroptosis.Nat Rev Mol Cell Biol. 2024 Jun;25(6):424-442. doi: 10.1038/s41580-024-00703-5. Epub 2024 Feb 16. Nat Rev Mol Cell Biol. 2024. PMID: 38366038 Review.
-
FSP1 inhibition enhances olaparib sensitivity in BRCA-proficient ovarian cancer patients via a nonferroptosis mechanism.Cell Death Differ. 2024 Apr;31(4):497-510. doi: 10.1038/s41418-024-01263-z. Epub 2024 Feb 19. Cell Death Differ. 2024. PMID: 38374229 Free PMC article.
-
Glycolysis-Related LINC02432/Hsa-miR-98-5p/HK2 Axis Inhibits Ferroptosis and Predicts Immune Infiltration, Tumor Mutation Burden, and Drug Sensitivity in Pancreatic Adenocarcinoma.Front Pharmacol. 2022 Jun 20;13:937413. doi: 10.3389/fphar.2022.937413. eCollection 2022. Front Pharmacol. 2022. PMID: 35795552 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

