mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron
- PMID: 35447072
- PMCID: PMC8947944
- DOI: 10.1016/j.cell.2022.03.038
mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron
Abstract
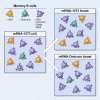
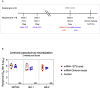
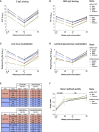
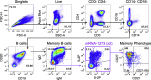
SARS-CoV-2 Omicron is highly transmissible and has substantial resistance to neutralization following immunization with ancestral spike-matched vaccines. It is unclear whether boosting with Omicron-matched vaccines would enhance protection. Here, nonhuman primates that received mRNA-1273 at weeks 0 and 4 were boosted at week 41 with mRNA-1273 or mRNA-Omicron. Neutralizing titers against D614G were 4,760 and 270 reciprocal ID50 at week 6 (peak) and week 41 (preboost), respectively, and 320 and 110 for Omicron. 2 weeks after the boost, titers against D614G and Omicron increased to 5,360 and 2,980 for mRNA-1273 boost and 2,670 and 1,930 for mRNA-Omicron, respectively. Similar increases against BA.2 were observed. Following either boost, 70%-80% of spike-specific B cells were cross-reactive against WA1 and Omicron. Equivalent control of virus replication in lower airways was observed following Omicron challenge 1 month after either boost. These data show that mRNA-1273 and mRNA-Omicron elicit comparable immunity and protection shortly after the boost.
Keywords: B cells; COVID-19; Omicron; SARS-CoV-2; T cells; antibody; boost; immune memory; mRNA vaccine; original antigenic sin.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests K.S.C. is an inventor on U.S. Patent no. 10,960,070 B2 and International Patent Application no. WO/2018/081318 entitled “Prefusion Coronavirus Spike Proteins and Their Use.” K.S.C. is an inventor on U.S. Patent Application no. 62/972,886 entitled “2019-nCoV Vaccine.” A.R.H., L.W., W.S., Y.Z., E.S.Y., J.R.M., P.D.K., M.R., N.J.S., and D.C.D. are inventors on U.S. Patent Application no. 63/147,419 entitled “Antibodies Targeting the Spike Protein of Coronaviruses.” L.P., A.V.R., B.N., D.V., A. Cook, A.D., K.S., H.A., and M.G.L. are employees of Bioqual. K.S.C., L.W., W.S., and Y.Z. are inventors on multiple U.S. Patent Applications entitled “Anti-Coronavirus Antibodies and Methods of Use.” G.-Y.C., G.S.-J., I.R., Y.-T.L., A.M., K.W., A. Carfi, and D.K.E. are employees of Moderna. M.S.S. serves on the scientific board of advisors for Moderna and Ocugen. The other authors declare no competing interests.
Figures










Similar articles
-
Ipsilateral or contralateral boosting of mice with mRNA vaccines confers equivalent immunity and protection against a SARS-CoV-2 Omicron strain.J Virol. 2024 Sep 17;98(9):e0057424. doi: 10.1128/jvi.00574-24. Epub 2024 Aug 28. J Virol. 2024. PMID: 39194250 Free PMC article.
-
Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant.Cell Rep Med. 2022 Jul 19;3(7):100679. doi: 10.1016/j.xcrm.2022.100679. Epub 2022 Jun 20. Cell Rep Med. 2022. PMID: 35798000 Free PMC article. Clinical Trial.
-
Vaccination and Omicron BA.1/BA.2 Convalescence Enhance Systemic but Not Mucosal Immunity against BA.4/5.Microbiol Spectr. 2023 Jun 15;11(3):e0516322. doi: 10.1128/spectrum.05163-22. Epub 2023 Apr 26. Microbiol Spectr. 2023. PMID: 37098903 Free PMC article.
-
Persistent T cell-mediated immune responses against Omicron variants after the third COVID-19 mRNA vaccine dose.Front Immunol. 2023 Jan 23;14:1099246. doi: 10.3389/fimmu.2023.1099246. eCollection 2023. Front Immunol. 2023. PMID: 36756112 Free PMC article.
-
Safety and immunogenicity against ancestral, Delta and Omicron virus variants following a booster dose of an inactivated whole-virus COVID-19 vaccine (VLA2001): Interim analysis of an open-label extension of the randomized, controlled, phase 3 COV-COMPARE trial.J Infect. 2023 Sep;87(3):242-254. doi: 10.1016/j.jinf.2023.06.022. Epub 2023 Jul 3. J Infect. 2023. PMID: 37406777 Clinical Trial.
Cited by
-
Comparative neutralization profiles of naive and breakthrough infections with Delta, Omicron BA.1 and BA.2 variants of SARS-CoV-2.Signal Transduct Target Ther. 2022 Sep 9;7(1):316. doi: 10.1038/s41392-022-01166-w. Signal Transduct Target Ther. 2022. PMID: 36085321 Free PMC article. No abstract available.
-
Delineating the Spread and Prevalence of SARS-CoV-2 Omicron Sublineages (BA.1-BA.5) and Deltacron Using Wastewater in the Western Cape, South Africa.J Infect Dis. 2022 Oct 17;226(8):1418-1427. doi: 10.1093/infdis/jiac356. J Infect Dis. 2022. PMID: 36017801 Free PMC article.
-
Do We Really Need Omicron Spike-Based Updated COVID-19 Vaccines? Evidence and Pipeline.Viruses. 2022 Nov 10;14(11):2488. doi: 10.3390/v14112488. Viruses. 2022. PMID: 36366586 Free PMC article. Review.
-
Mucosal adenovirus vaccine boosting elicits IgA and durably prevents XBB.1.16 infection in nonhuman primates.Nat Immunol. 2024 Oct;25(10):1913-1927. doi: 10.1038/s41590-024-01951-5. Epub 2024 Sep 3. Nat Immunol. 2024. PMID: 39227514 Free PMC article.
-
The potential of Beta variant containing COVID booster vaccines for chasing Omicron in 2022.Nat Commun. 2022 Oct 2;13(1):5794. doi: 10.1038/s41467-022-33549-6. Nat Commun. 2022. PMID: 36184631 Free PMC article.
References
-
- Abdullah F., Myers J., Basu D., Tintinger G., Ueckermann V., Mathebula M., Ramlall R., Spoor S., de Villiers T., Van der Walt Z., et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, South Africa. Int. J. Infect. Dis. 2022;116:38–42. - PMC - PubMed
-
- Andrews N., Stowe J., Kirsebom F., Toffa S., Sachdeva R., Gower C., Ramsay M., Lopez Bernal J.L. Effectiveness of COVID-19 booster vaccines against covid-19-related symptoms, hospitalisation and death in England. Nat. Med. Published online January. 2022;14:2022. doi: 10.1038/s41591-022-01699-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

