Evolutionarily conserved properties of CLCA proteins 1, 3 and 4, as revealed by phylogenetic and biochemical studies in avian homologues
- PMID: 35417490
- PMCID: PMC9007345
- DOI: 10.1371/journal.pone.0266937
Evolutionarily conserved properties of CLCA proteins 1, 3 and 4, as revealed by phylogenetic and biochemical studies in avian homologues
Abstract
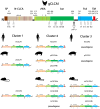
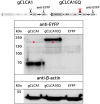
Species-specific diversities are particular features of mammalian chloride channel regulator, calcium activated (CLCA) genes. In contrast to four complex gene clusters in mammals, only two CLCA genes appear to exist in chickens. CLCA2 is conserved in both, while only the galline CLCA1 (gCLCA1) displays close genetic distance to mammalian clusters 1, 3 and 4. In this study, sequence analyses and biochemical characterizations revealed that gCLCA1 as a putative avian prototype shares common protein domains and processing features with all mammalian CLCA homologues. It has a transmembrane (TM) domain in the carboxy terminal region and its mRNA and protein were detected in the alimentary canal, where the protein was localized in the apical membrane of enterocytes, similar to CLCA4. Both mammals and birds seem to have at least one TM domain containing CLCA protein with complex glycosylation in the apical membrane of enterocytes. However, some characteristic features of mammalian CLCA1 and 3 including entire protein secretion and expression in cell types other than enterocytes seem to be dispensable for chicken. Phylogenetic analyses including twelve bird species revealed that avian CLCA1 and mammalian CLCA3 form clades separate from a major branch containing mammalian CLCA1 and 4. Overall, our data suggest that gCLCA1 and mammalian CLCA clusters 1, 3 and 4 stem from a common ancestor which underwent complex gene diversification in mammals but not in birds.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
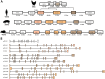
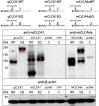
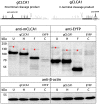
 -icons. Numbers display the absolute position, for protein domains further specified by the first or last three amino acids of each domain, respectively.
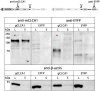
-icons. Numbers display the absolute position, for protein domains further specified by the first or last three amino acids of each domain, respectively.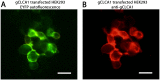



Similar articles
-
Genomic, biochemical and expressional properties reveal strong conservation of the CLCA2 gene in birds and mammals.PeerJ. 2022 Nov 8;10:e14202. doi: 10.7717/peerj.14202. eCollection 2022. PeerJ. 2022. PMID: 36389428 Free PMC article.
-
Impaired autoproteolytic cleavage of mCLCA6, a murine integral membrane protein expressed in enterocytes, leads to cleavage at the plasma membrane instead of the endoplasmic reticulum.Mol Cells. 2012 Mar;33(3):251-7. doi: 10.1007/s10059-012-2217-1. Epub 2012 Feb 15. Mol Cells. 2012. PMID: 22350745 Free PMC article.
-
Expression analysis of the CLCA gene family in mouse and human with emphasis on the nervous system.BMC Dev Biol. 2009 Feb 11;9:10. doi: 10.1186/1471-213X-9-10. BMC Dev Biol. 2009. PMID: 19210762 Free PMC article.
-
The CLCA gene family: putative therapeutic target for respiratory diseases.Inflamm Allergy Drug Targets. 2009 Jun;8(2):146-60. doi: 10.2174/187152809788462590. Inflamm Allergy Drug Targets. 2009. PMID: 19530997 Review.
-
Structure and function of CLCA proteins.Physiol Rev. 2005 Jul;85(3):1061-92. doi: 10.1152/physrev.00016.2004. Physiol Rev. 2005. PMID: 15987802 Review.
Cited by
-
Self-Cleavage of Human Chloride Channel Accessory 2 Causes a Conformational Shift That Depends on Membrane Anchorage and Is Required for Its Regulation of Store-Operated Calcium Entry.Biomedicines. 2023 Oct 28;11(11):2915. doi: 10.3390/biomedicines11112915. Biomedicines. 2023. PMID: 38001916 Free PMC article.
-
Genomic, biochemical and expressional properties reveal strong conservation of the CLCA2 gene in birds and mammals.PeerJ. 2022 Nov 8;10:e14202. doi: 10.7717/peerj.14202. eCollection 2022. PeerJ. 2022. PMID: 36389428 Free PMC article.
References
-
- Hoshino M, Morita S, Iwashita H, Sagiya Y, Nagi T, Nakanishi A, et al.. Increased expression of the human Ca2+-activated Cl− channel 1 (CaCC1) gene in the asthmatic airway. American journal of respiratory and critical care medicine. 2002;165(8):1132–6. doi: 10.1164/ajrccm.165.8.2107068 - DOI - PubMed
-
- Hauber HP, Manoukian JJ, Nguyen LH, Sobol SE, Levitt RC, Holroyd KJ, et al.. Increased expression of interleukin-9, interleukin-9 receptor, and the calcium-activated chloride channel hCLCA1 in the upper airways of patients with cystic fibrosis. The Laryngoscope. 2003;113(6):1037–42. doi: 10.1097/00005537-200306000-00022 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

