Congenital microtia patients: the genetically engineered exosomes released from porous gelatin methacryloyl hydrogel for downstream small RNA profiling, functional modulation of microtia chondrocytes and tissue-engineered ear cartilage regeneration
- PMID: 35346221
- PMCID: PMC8962601
- DOI: 10.1186/s12951-022-01352-6
Congenital microtia patients: the genetically engineered exosomes released from porous gelatin methacryloyl hydrogel for downstream small RNA profiling, functional modulation of microtia chondrocytes and tissue-engineered ear cartilage regeneration
Abstract
Background: Mesenchymal stem cells (MSCs) exosomes were previously shown to be effective in articular cartilage repairing. However, whether MSCs exosomes promote mature cartilage formation of microtia chondrocytes and the underlying mechanism of action remains unknown. Additionally, some hurdles, such as the low yield and unsatisfactory therapeutic effects of natural exosomes have emerged when considering the translation of exosomes-therapeutics to clinical practices or industrial production. Herein, we investigated the roles of human adipose-derived stem cells (ADSCs) exosomes in modulating microtia chondrocytes and the underlying mechanism of action. Special attention was also paid to the mass production and functional modification of ADSCs exosomes.
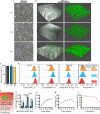
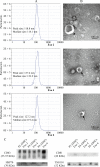
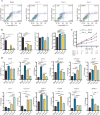
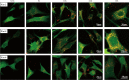
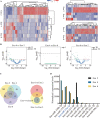
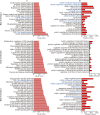
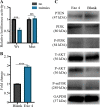
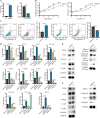
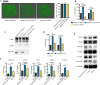
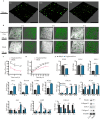
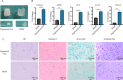
Results: We firstly used porous gelatin methacryloyl (Porous Gelma) hydrogel with pores size of 100 to 200 μm for 3D culture of passage 2, 4 and 6 ADSCs (P2, P4 and P6 ADSCs, respectively), and obtained their corresponding exosomes (Exo 2, Exo 4 and Exo 6, respectively). In vitro results showed Exo 2 outperformed both Exo 4 and Exo 6 in enhancing cell proliferation and attenuating apoptosis. However, both Exo 4 and Exo 6 promoted chondrogenesis more than Exo 2 did. Small RNA sequencing results indicated Exo 4 was similar to Exo 6 in small RNA profiles and consistently upregulated PI3K/AKT/mTOR signaling pathway. Notably, we found hsa-miR-23a-3p was highly expressed in Exo 4 and Exo 6 compared to Exo 2, and they modulated microtia chondrocytes by transferring hsa-miR-23a-3p to suppress PTEN expression, and consequently to activate PI3K/AKT/mTOR signaling pathway. Then, we designed genetically engineered exosomes by directly transfecting agomir-23a-3p into parent P4 ADSCs and isolated hsa-miR-23a-3p-rich exosomes for optimizing favorable effects on cell viability and new cartilage formation. Subsequently, we applied the engineered exosomes to in vitro and in vivo tissue-engineered cartilage culture and consistently found that the engineered exosomes could enhance cell proliferation, attenuate apoptosis and promote cartilage regeneration.
Conclusions: Taken together, the porous Gelma hydrogel could be applied to exosomes mass production, and functional modification could be achieved by selecting P4 ADSCs as parent cells and genetically modifying ADSCs. Our engineered exosomes are a promising candidate for tissue-engineered ear cartilage regeneration.
Keywords: Adipose-derived stem cells; Engineered exosomes; Microtia; Porous gelatin methacryloyl; Tissue-engineered ear cartilage.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Engineered hsa-miR-455-3p-Abundant Extracellular Vesicles Derived from 3D-Cultured Adipose Mesenchymal Stem Cells for Tissue-Engineering Hyaline Cartilage Regeneration.Adv Healthc Mater. 2024 Jul;13(18):e2304194. doi: 10.1002/adhm.202304194. Epub 2024 Apr 3. Adv Healthc Mater. 2024. PMID: 38508211
-
Co-culture of RhoA-overexpressed microtia chondrocytes and adipose-derived stem cells in the construction of tissue-engineered ear-shaped cartilage.Stem Cells. 2024 Jun 14;42(6):554-566. doi: 10.1093/stmcls/sxae026. Stem Cells. 2024. PMID: 38613477
-
miR-23a-3p-abundant small extracellular vesicles released from Gelma/nanoclay hydrogel for cartilage regeneration.J Extracell Vesicles. 2020 Jun 16;9(1):1778883. doi: 10.1080/20013078.2020.1778883. J Extracell Vesicles. 2020. PMID: 32939233 Free PMC article.
-
Exosomes loaded with chondrogenic stimuli agents combined with 3D bioprinting hydrogel in the treatment of osteoarthritis and cartilage degeneration.Biomed Pharmacother. 2023 Dec;168:115715. doi: 10.1016/j.biopha.2023.115715. Epub 2023 Oct 17. Biomed Pharmacother. 2023. PMID: 37857246 Review.
-
The therapeutic potential of stem cell-derived exosomes in the ulcerative colitis and colorectal cancer.Stem Cell Res Ther. 2022 Apr 1;13(1):138. doi: 10.1186/s13287-022-02811-5. Stem Cell Res Ther. 2022. PMID: 35365226 Free PMC article. Review.
Cited by
-
[Research progress of three-dimensional bioprinting technology in auricle repair and reconstruction].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2024 Jun 15;38(6):763-768. doi: 10.7507/1002-1892.202403001. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2024. PMID: 38918200 Free PMC article. Review. Chinese.
-
Huc-MSC-derived exosomes modified with the targeting peptide of aHSCs for liver fibrosis therapy.J Nanobiotechnology. 2022 Oct 1;20(1):432. doi: 10.1186/s12951-022-01636-x. J Nanobiotechnology. 2022. PMID: 36183106 Free PMC article.
-
Advanced Hybrid Strategies of GelMA Composite Hydrogels in Bone Defect Repair.Polymers (Basel). 2024 Oct 29;16(21):3039. doi: 10.3390/polym16213039. Polymers (Basel). 2024. PMID: 39518248 Free PMC article. Review.
-
Signaling pathways activated and regulated by stem cell-derived exosome therapy.Cell Biosci. 2024 Aug 20;14(1):105. doi: 10.1186/s13578-024-01277-7. Cell Biosci. 2024. PMID: 39164778 Free PMC article. Review.
-
Engineering of MSC-Derived Exosomes: A Promising Cell-Free Therapy for Osteoarthritis.Membranes (Basel). 2022 Jul 28;12(8):739. doi: 10.3390/membranes12080739. Membranes (Basel). 2022. PMID: 36005656 Free PMC article. Review.
References
-
- Landau S, Szklanny AA, Machour M, Kaplan B, Shandalov Y, Redenski I, et al. Human-engineered auricular reconstruction (hEAR) by 3D-printed molding with human-derived auricular and costal chondrocytes and adipose-derived mesenchymal stem cells. Biofabrication. 2021;14:1. - PubMed
-
- Chen X, Zhang R, Zhang Q, Xu Z, Xu F, Li D, Li Y. Microtia patients: auricular chondrocyte ECM is promoted by CGF through IGF-1 activation of the IGF-1R/PI3K/AKT pathway. J Cell Physiol. 2019;234:21817–24. - PubMed
-
- Nakao H, Jacquet RD, Shasti M, Isogai N, Murthy AS, Landis WJ. Long-term comparison between human normal conchal and microtia chondrocytes regenerated by tissue engineering on nanofiber polyglycolic acid scaffolds. Plast Reconstr Surg. 2017;139:911e–921e. - PubMed
-
- Zhang L, He A, Yin Z, Yu Z, Luo X, Liu W, et al. Regeneration of human-ear-shaped cartilage by co-culturing human microtia chondrocytes with BMSCs. Biomaterials. 2014;35:4878–87. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

