Homer1 promotes the conversion of A1 astrocytes to A2 astrocytes and improves the recovery of transgenic mice after intracerebral hemorrhage
- PMID: 35287697
- PMCID: PMC8922810
- DOI: 10.1186/s12974-022-02428-8
Homer1 promotes the conversion of A1 astrocytes to A2 astrocytes and improves the recovery of transgenic mice after intracerebral hemorrhage
Abstract
Background: Inflammation induced by intracerebral hemorrhage (ICH) is one of the main causes of the high mortality and poor prognosis of patients with ICH. A1 astrocytes are closely associated with neuroinflammation and neurotoxicity, whereas A2 astrocytes are neuroprotective. Homer scaffolding protein 1 (Homer1) plays a protective role in ischemic encephalopathy and neurodegenerative diseases. However, the role of Homer1 in ICH-induced inflammation and the effect of Homer1 on the phenotypic conversion of astrocytes remain unknown.
Methods: Femoral artery autologous blood from C57BL/6 mice was used to create an ICH model. We use the A1 phenotype marker C3 and A2 phenotype marker S100A10 to detect astrocyte conversion after ICH. Homer1 overexpression/knock-down mice were constructed by adeno-associated virus (AAV) infection to explore the role of Homer1 and its mechanism of action after ICH. Finally, Homer1 protein and selumetinib were injected into in situ hemorrhage sites in the brains of Homer1flox/flox/Nestin-Cre+/- mice to study the efficacy of Homer1 in the treatment of ICH by using a mouse cytokine array to explore the potential mechanism.
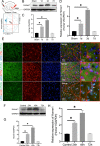
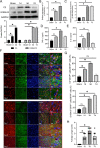
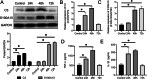
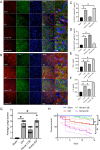
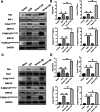
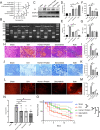
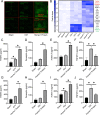
Results: The expression of Homer1 peaked on the third day after ICH and colocalized with astrocytes. Homer1 promotes A1 phenotypic conversion in astrocytes in vivo and in vitro. Overexpression of Homer1 inhibits the activation of MAPK signaling, whereas Homer1 knock-down increases the expression of pathway-related proteins. The Homer1 protein and selumetinib, a non-ATP competitive MEK1/2 inhibitor, improved the outcome in ICH in Homer1flox/flox/Nestin-Cre+/- mice. The efficacy of Homer1 in the treatment of ICH is associated with reduced expression of the inflammatory factor TNFSF10 and increased expression of the anti-inflammatory factors activin A, persephin, and TWEAK.
Conclusions: Homer1 plays an important role in inhibiting inflammation after ICH by suppressing the A1 phenotype conversion in astrocytes. In situ injection of Homer1 protein may be a novel and effective method for the treatment of inflammation after ICH.
Keywords: Astrocytes; Homer1; Inflammation; Intracerebral hemorrhage; Phenotype.
© 2022. The Author(s).
Conflict of interest statement
The authors declared that they have no competing interests.
Figures

Similar articles
-
Extracellular vesicle encapsulated Homer1a as novel nanotherapeutics against intracerebral hemorrhage in a mouse model.J Neuroinflammation. 2024 Apr 6;21(1):85. doi: 10.1186/s12974-024-03088-6. J Neuroinflammation. 2024. PMID: 38582897 Free PMC article.
-
Homer1 ameliorates ischemic stroke by inhibiting necroptosis-induced neuronal damage and neuroinflammation.Inflamm Res. 2024 Jan;73(1):131-144. doi: 10.1007/s00011-023-01824-x. Epub 2023 Dec 13. Inflamm Res. 2024. PMID: 38091015 Free PMC article.
-
Activation of chaperone-mediated autophagy exerting neuroprotection effect on intracerebral hemorrhage-induced neuronal injury by targeting Lamp2a.Exp Neurol. 2024 Dec;382:114986. doi: 10.1016/j.expneurol.2024.114986. Epub 2024 Oct 4. Exp Neurol. 2024. PMID: 39368534
-
Astrocytes in intracerebral hemorrhage: impact and therapeutic objectives.Front Mol Neurosci. 2024 Feb 14;17:1327472. doi: 10.3389/fnmol.2024.1327472. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38419793 Free PMC article. Review.
-
The hallmark and crosstalk of immune cells after intracerebral hemorrhage: Immunotherapy perspectives.Front Neurosci. 2023 Jan 12;16:1117999. doi: 10.3389/fnins.2022.1117999. eCollection 2022. Front Neurosci. 2023. PMID: 36711145 Free PMC article. Review.
Cited by
-
Transformation of A1/A2 Astrocytes Participates in Brain Ischemic Tolerance Induced by Cerebral Ischemic Preconditioning via Inhibiting NDRG2.Neurochem Res. 2024 Jul;49(7):1665-1676. doi: 10.1007/s11064-024-04134-8. Epub 2024 Feb 27. Neurochem Res. 2024. PMID: 38411782
-
Glial cells: an important switch for the vascular function of the central nervous system.Front Cell Neurosci. 2023 May 3;17:1166770. doi: 10.3389/fncel.2023.1166770. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37206667 Free PMC article. Review.
-
Extracellular vesicle encapsulated Homer1a as novel nanotherapeutics against intracerebral hemorrhage in a mouse model.J Neuroinflammation. 2024 Apr 6;21(1):85. doi: 10.1186/s12974-024-03088-6. J Neuroinflammation. 2024. PMID: 38582897 Free PMC article.
-
Homer1 Protects against Retinal Ganglion Cell Pyroptosis by Inhibiting Endoplasmic Reticulum Stress-Associated TXNIP/NLRP3 Inflammasome Activation after Middle Cerebral Artery Occlusion-Induced Retinal Ischemia.Int J Mol Sci. 2023 Nov 27;24(23):16811. doi: 10.3390/ijms242316811. Int J Mol Sci. 2023. PMID: 38069134 Free PMC article.
-
Homer1a reduces inflammatory response after retinal ischemia/reperfusion injury.Neural Regen Res. 2024 Jul 1;19(7):1608-1617. doi: 10.4103/1673-5374.386490. Epub 2023 Nov 8. Neural Regen Res. 2024. PMID: 38051906 Free PMC article.
References
-
- Spagnuolo M, et al. Brain-derived neurotrophic factor modulates cholesterol homeostasis and Apolipoprotein E synthesis in human cell models of astrocytes and neurons. J Cell Physiol. 2018;233(9):6925–6943. - PubMed
-
- Bijelić D, et al. Central nervous system-infiltrated immune cells induce calcium increase in astrocytes via astroglial purinergic signaling. J Neurosci Res. 2020;98(11):2317–2332. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

