What's in a Gene? The Outstanding Diversity of MAPT
- PMID: 35269461
- PMCID: PMC8909800
- DOI: 10.3390/cells11050840
What's in a Gene? The Outstanding Diversity of MAPT
Abstract
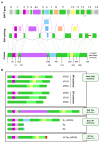
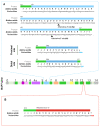
Tau protein is a microtubule-associated protein encoded by the MAPT gene that carries out a myriad of physiological functions and has been linked to certain pathologies collectively termed tauopathies, including Alzheimer's disease, frontotemporal dementia, Huntington's disease, progressive supranuclear palsy, etc. Alternative splicing is a physiological process by which cells generate several transcripts from one single gene and may in turn give rise to different proteins from the same gene. MAPT transcripts have been proven to be subjected to alternative splicing, generating six main isoforms in the central nervous system. Research throughout the years has demonstrated that the splicing landscape of the MAPT gene is far more complex than that, including at least exon skipping events, the use of 3' and 5' alternative splice sites and, as has been recently discovered, also intron retention. In addition, MAPT alternative splicing has been showed to be regulated spatially and developmentally, further evidencing the complexity of the gene's splicing regulation. It is unclear what would drive the need for the existence of so many isoforms encoded by the same gene, but a wide range of functions have been ascribed to these Tau isoforms, both in physiology and pathology. In this review we offer a comprehensive up-to-date exploration of the mechanisms leading to the outstanding diversity of isoforms expressed from the MAPT gene and the functions in which such isoforms are involved, including their potential role in the onset and development of tauopathies such as Alzheimer's disease.
Keywords: Alzheimer’s disease; MAPT; Tau protein; alternative splicing; intron retention.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Tau Isoforms: Gaining Insight into MAPT Alternative Splicing.Int J Mol Sci. 2022 Dec 6;23(23):15383. doi: 10.3390/ijms232315383. Int J Mol Sci. 2022. PMID: 36499709 Free PMC article. Review.
-
Intron retention as a productive mechanism in human MAPT: RNA species generated by retention of intron 3.EBioMedicine. 2024 Feb;100:104953. doi: 10.1016/j.ebiom.2023.104953. Epub 2024 Jan 5. EBioMedicine. 2024. PMID: 38181704 Free PMC article.
-
Tau alternative splicing in familial and sporadic tauopathies.Biochem Soc Trans. 2012 Aug;40(4):677-80. doi: 10.1042/BST20120091. Biochem Soc Trans. 2012. PMID: 22817715 Review.
-
Dysregulated coordination of MAPT exon 2 and exon 10 splicing underlies different tau pathologies in PSP and AD.Acta Neuropathol. 2022 Feb;143(2):225-243. doi: 10.1007/s00401-021-02392-2. Epub 2021 Dec 7. Acta Neuropathol. 2022. PMID: 34874463 Free PMC article.
-
The Role of Tau Proteoforms in Health and Disease.Mol Neurobiol. 2023 Sep;60(9):5155-5166. doi: 10.1007/s12035-023-03387-8. Epub 2023 Jun 2. Mol Neurobiol. 2023. PMID: 37266762 Review.
Cited by
-
Psychosis in Parkinson's Disease: A Lesson from Genetics.Genes (Basel). 2022 Jun 20;13(6):1099. doi: 10.3390/genes13061099. Genes (Basel). 2022. PMID: 35741861 Free PMC article. Review.
-
Tau Isoforms: Gaining Insight into MAPT Alternative Splicing.Int J Mol Sci. 2022 Dec 6;23(23):15383. doi: 10.3390/ijms232315383. Int J Mol Sci. 2022. PMID: 36499709 Free PMC article. Review.
-
A novel MAPT variant (E342K) as a cause of familial progressive supranuclear palsy.Front Neurol. 2024 Apr 19;15:1372507. doi: 10.3389/fneur.2024.1372507. eCollection 2024. Front Neurol. 2024. PMID: 38708005 Free PMC article.
-
Regulation of Tau Expression in Superior Cervical Ganglion (SCG) Neurons In Vivo and In Vitro.Cells. 2023 Jan 5;12(2):226. doi: 10.3390/cells12020226. Cells. 2023. PMID: 36672160 Free PMC article.
-
Evolutionary perspective of Big tau structure: 4a exon variants of MAPT.Front Mol Neurosci. 2022 Dec 2;15:1019999. doi: 10.3389/fnmol.2022.1019999. eCollection 2022. Front Mol Neurosci. 2022. PMID: 36533137 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

