Arabidopsis RBV is a conserved WD40 repeat protein that promotes microRNA biogenesis and ARGONAUTE1 loading
- PMID: 35260568
- PMCID: PMC8904849
- DOI: 10.1038/s41467-022-28872-x
Arabidopsis RBV is a conserved WD40 repeat protein that promotes microRNA biogenesis and ARGONAUTE1 loading
Abstract
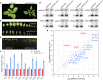
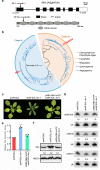
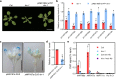
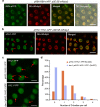
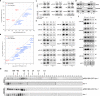
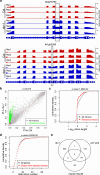
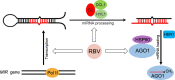
MicroRNAs (miRNAs) play crucial roles in gene expression regulation through RNA cleavage or translation repression. Here, we report the identification of an evolutionarily conserved WD40 domain protein as a player in miRNA biogenesis in Arabidopsis thaliana. A mutation in the REDUCTION IN BLEACHED VEIN AREA (RBV) gene encoding a WD40 domain protein led to the suppression of leaf bleaching caused by an artificial miRNA; the mutation also led to a global reduction in the accumulation of endogenous miRNAs. The nuclear protein RBV promotes the transcription of MIR genes into pri-miRNAs by enhancing the occupancy of RNA polymerase II (Pol II) at MIR gene promoters. RBV also promotes the loading of miRNAs into AGO1. In addition, RNA-seq revealed a global splicing defect in the mutant. Thus, this evolutionarily conserved, nuclear WD40 domain protein acts in miRNA biogenesis and RNA splicing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
TRANSPORTIN1 Promotes the Association of MicroRNA with ARGONAUTE1 in Arabidopsis.Plant Cell. 2016 Oct;28(10):2576-2585. doi: 10.1105/tpc.16.00384. Epub 2016 Sep 23. Plant Cell. 2016. PMID: 27662897 Free PMC article.
-
HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis.Plant J. 2015 Mar;81(6):861-70. doi: 10.1111/tpj.12772. Epub 2015 Feb 19. Plant J. 2015. PMID: 25619693 Free PMC article.
-
The N-terminal extension of Arabidopsis ARGONAUTE 1 is essential for microRNA activities.PLoS Genet. 2023 Mar 8;19(3):e1010450. doi: 10.1371/journal.pgen.1010450. eCollection 2023 Mar. PLoS Genet. 2023. PMID: 36888599 Free PMC article.
-
Novel Nuclear Functions of Arabidopsis ARGONAUTE1: Beyond RNA Interference.Plant Physiol. 2019 Mar;179(3):1030-1039. doi: 10.1104/pp.18.01351. Epub 2019 Jan 3. Plant Physiol. 2019. PMID: 30606888 Free PMC article. Review.
-
Biogenesis, functions and fate of plant microRNAs.J Cell Physiol. 2012 Sep;227(9):3163-8. doi: 10.1002/jcp.24052. J Cell Physiol. 2012. PMID: 22252306 Review.
Cited by
-
The Intersection of Non-Coding RNAs Contributes to Forest Trees' Response to Abiotic Stress.Int J Mol Sci. 2022 Jun 7;23(12):6365. doi: 10.3390/ijms23126365. Int J Mol Sci. 2022. PMID: 35742808 Free PMC article. Review.
-
The spliceosome-associated protein CWC15 promotes miRNA biogenesis in Arabidopsis.Nat Commun. 2024 Mar 16;15(1):2399. doi: 10.1038/s41467-024-46676-z. Nat Commun. 2024. PMID: 38493158 Free PMC article.
-
Arabidopsis AAR2, a conserved splicing factor in eukaryotes, acts in microRNA biogenesis.Proc Natl Acad Sci U S A. 2022 Oct 11;119(41):e2208415119. doi: 10.1073/pnas.2208415119. Epub 2022 Oct 3. Proc Natl Acad Sci U S A. 2022. PMID: 36191209 Free PMC article.
-
Peptidyl-prolyl isomerase Cyclophilin71 promotes SERRATE phase separation and miRNA processing in Arabidopsis.Proc Natl Acad Sci U S A. 2023 Sep 5;120(36):e2305244120. doi: 10.1073/pnas.2305244120. Epub 2023 Aug 28. Proc Natl Acad Sci U S A. 2023. PMID: 37639607 Free PMC article.
-
microRNA biogenesis and stabilization in plants.Fundam Res. 2023 Mar 17;3(5):707-717. doi: 10.1016/j.fmre.2023.02.023. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933298 Free PMC article. Review.
References
-
- Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019;70:489–525. - PubMed

