Epigenomic priming of immune genes implicates oligodendroglia in multiple sclerosis susceptibility
- PMID: 35093191
- PMCID: PMC9810341
- DOI: 10.1016/j.neuron.2021.12.034
Epigenomic priming of immune genes implicates oligodendroglia in multiple sclerosis susceptibility
Abstract
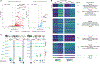
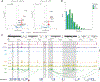
Multiple sclerosis (MS) is characterized by a targeted attack on oligodendroglia (OLG) and myelin by immune cells, which are thought to be the main drivers of MS susceptibility. We found that immune genes exhibit a primed chromatin state in single mouse and human OLG in a non-disease context, compatible with transitions to immune-competent states in MS. We identified BACH1 and STAT1 as transcription factors involved in immune gene regulation in oligodendrocyte precursor cells (OPCs). A subset of immune genes presents bivalency of H3K4me3/H3K27me3 in OPCs, with Polycomb inhibition leading to their increased activation upon interferon gamma (IFN-γ) treatment. Some MS susceptibility single-nucleotide polymorphisms (SNPs) overlap with these regulatory regions in mouse and human OLG. Treatment of mouse OPCs with IFN-γ leads to chromatin architecture remodeling at these loci and altered expression of interacting genes. Thus, the susceptibility for MS may involve OLG, which therefore constitutes novel targets for immunological-based therapies for MS.
Keywords: Polycomb; chromatin; genome-wide association studies; histone modifications; major histocompatibility complex; multiple sclerosis; myelin; neuroimmunology; oligodendrocyte; single-nucleotide polymorphisms.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests H.Y.C. is a co-founder of Accent Therapeutics and Boundless Bio and an advisor to 10x Genomics, Arsenal Biosciences, and Spring Discovery. D.M. and D.C. are employees at Roche Pharma Research and Early Development. The other authors declare no competing interests.
Figures




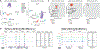

Comment in
-
Primed and ready.Nat Rev Neurosci. 2022 Apr;23(4):189. doi: 10.1038/s41583-022-00575-8. Nat Rev Neurosci. 2022. PMID: 35194224 No abstract available.
-
Immune genes outside immune cells for multiple sclerosis.Neuron. 2022 Apr 6;110(7):1090-1092. doi: 10.1016/j.neuron.2022.03.017. Neuron. 2022. PMID: 35390286
Similar articles
-
Histamine Receptor 3 negatively regulates oligodendrocyte differentiation and remyelination.PLoS One. 2017 Dec 18;12(12):e0189380. doi: 10.1371/journal.pone.0189380. eCollection 2017. PLoS One. 2017. PMID: 29253893 Free PMC article.
-
Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis.Nat Med. 2018 Dec;24(12):1837-1844. doi: 10.1038/s41591-018-0236-y. Epub 2018 Nov 12. Nat Med. 2018. PMID: 30420755 Free PMC article.
-
Immune genes outside immune cells for multiple sclerosis.Neuron. 2022 Apr 6;110(7):1090-1092. doi: 10.1016/j.neuron.2022.03.017. Neuron. 2022. PMID: 35390286
-
MicroRNAs in oligodendrocyte development and remyelination.J Neurochem. 2022 Aug;162(4):310-321. doi: 10.1111/jnc.15618. Epub 2022 May 10. J Neurochem. 2022. PMID: 35536759 Review.
-
Interferon regulatory factor 1 regulation of oligodendrocyte injury and inflammatory demyelination.Rev Neurosci. 2012 Jan 26;23(2):145-52. doi: 10.1515/revneuro-2011-068. Rev Neurosci. 2012. PMID: 22499673 Review.
Cited by
-
Histone methylation in Epstein-Barr virus-associated diseases.Epigenomics. 2024;16(11-12):865-877. doi: 10.1080/17501911.2024.2345040. Epub 2024 May 10. Epigenomics. 2024. PMID: 38869454 Review.
-
MHC class I and MHC class II reporter mice enable analysis of immune oligodendroglia in mouse models of multiple sclerosis.Elife. 2023 Apr 14;12:e82938. doi: 10.7554/eLife.82938. Elife. 2023. PMID: 37057892 Free PMC article.
-
Tissue-specific enhancer-gene maps from multimodal single-cell data identify causal disease alleles.Nat Genet. 2024 Apr;56(4):615-626. doi: 10.1038/s41588-024-01682-1. Epub 2024 Apr 9. Nat Genet. 2024. PMID: 38594305 Free PMC article.
-
Cellular Therapy in Experimental Autoimmune Encephalomyelitis as an Adjuvant Treatment to Translate for Multiple Sclerosis.Int J Mol Sci. 2024 Jun 26;25(13):6996. doi: 10.3390/ijms25136996. Int J Mol Sci. 2024. PMID: 39000105 Free PMC article.
-
Transcriptional abnormalities in induced pluripotent stem cell-derived oligodendrocytes of individuals with primary progressive multiple sclerosis.Front Cell Neurosci. 2022 Sep 28;16:972144. doi: 10.3389/fncel.2022.972144. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36246526 Free PMC article.
References
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

