Osteoblastic Swedish mutant APP expedites brain deficits by inducing endoplasmic reticulum stress-driven senescence
- PMID: 34824365
- PMCID: PMC8617160
- DOI: 10.1038/s42003-021-02843-2
Osteoblastic Swedish mutant APP expedites brain deficits by inducing endoplasmic reticulum stress-driven senescence
Abstract
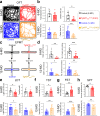
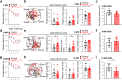
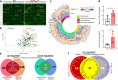
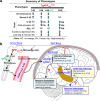
Patients with Alzheimer's disease (AD) often have osteoporosis or osteopenia. However, their direct link and relationship remain largely unclear. Previous studies have detected osteoporotic deficits in young adult Tg2576 and TgAPPsweOCN mice, which express APPswe (Swedish mutant) ubiquitously and selectively in osteoblast (OB)-lineage cells. This raises the question, whether osteoblastic APPswe contributes to AD development. Here, we provide evidence that TgAPPsweOCN mice also exhibit AD-relevant brain pathologies and behavior phenotypes. Some brain pathologies include age-dependent and regional-selective increases in glial activation and pro-inflammatory cytokines, which are accompanied by behavioral phenotypes such as anxiety, depression, and altered learning and memory. Further cellular studies suggest that APPswe, but not APPwt or APPlon (London mutant), in OB-lineage cells induces endoplasmic reticulum-stress driven senescence, driving systemic and cortex inflammation as well as behavioral changes in 6-month-old TgAPPsweOCN mice. These results therefore reveal an unrecognized function of osteoblastic APPswe to brain axis in AD development.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Swedish mutant APP suppresses osteoblast differentiation and causes osteoporotic deficit, which are ameliorated by N-acetyl-L-cysteine.J Bone Miner Res. 2013 Oct;28(10):2122-35. doi: 10.1002/jbmr.1954. J Bone Miner Res. 2013. PMID: 23649480 Free PMC article.
-
Muscular Swedish mutant APP-to-Brain axis in the development of Alzheimer's disease.Cell Death Dis. 2022 Nov 10;13(11):952. doi: 10.1038/s41419-022-05378-4. Cell Death Dis. 2022. PMID: 36357367 Free PMC article.
-
APP promotes osteoblast survival and bone formation by regulating mitochondrial function and preventing oxidative stress.Cell Death Dis. 2018 Oct 22;9(11):1077. doi: 10.1038/s41419-018-1123-7. Cell Death Dis. 2018. PMID: 30349052 Free PMC article.
-
Comparative studies using the Morris water maze to assess spatial memory deficits in two transgenic mouse models of Alzheimer's disease.Clin Exp Pharmacol Physiol. 2014 Oct;41(10):798-806. doi: 10.1111/1440-1681.12277. Clin Exp Pharmacol Physiol. 2014. PMID: 25115283
-
Effects of accelerated senescence on learning and memory, locomotion and anxiety-like behavior in APP/PS1 mouse model of Alzheimer's disease.J Neurol Sci. 2013 Dec 15;335(1-2):145-54. doi: 10.1016/j.jns.2013.09.018. Epub 2013 Sep 21. J Neurol Sci. 2013. PMID: 24095271
Cited by
-
Elevated PDGF-BB from Bone Impairs Hippocampal Vasculature by Inducing PDGFRβ Shedding from Pericytes.Adv Sci (Weinh). 2023 Jul;10(20):e2206938. doi: 10.1002/advs.202206938. Epub 2023 Apr 27. Adv Sci (Weinh). 2023. PMID: 37102631 Free PMC article.
-
Advances in the cell biology of the trafficking and processing of amyloid precursor protein: impact of familial Alzheimer's disease mutations.Biochem J. 2024 Oct 2;481(19):1297-1325. doi: 10.1042/BCJ20240056. Biochem J. 2024. PMID: 39302110 Free PMC article. Review.
-
Attenuation of Alzheimer's brain pathology in 5XFAD mice by PTH1-34, a peptide of parathyroid hormone.Alzheimers Res Ther. 2023 Mar 14;15(1):53. doi: 10.1186/s13195-023-01202-z. Alzheimers Res Ther. 2023. PMID: 36918976 Free PMC article.
-
Network Analysis of Brain and Bone Tissue Transcripts Reveals Shared Molecular Mechanisms Underlying Alzheimer's Disease and Related Dementias and Osteoporosis.J Gerontol A Biol Sci Med Sci. 2024 Nov 1;79(11):glae211. doi: 10.1093/gerona/glae211. J Gerontol A Biol Sci Med Sci. 2024. PMID: 39194133
References
-
- Leng, F. & Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat. Rev. Neurol.17, 157–172 (2020). - PubMed
-
- Wang J, Gu BJ, Masters CL, Wang YJ. A systemic view of Alzheimer disease—insights from amyloid-beta metabolism beyond the brain. Nat. Rev. Neurol. 2017;13:612–623. - PubMed
-
- Mjoberg B, Hellquist E, Mallmin H, Lindh U. Aluminum, Alzheimer’s disease and bone fragility. Acta Orthopaedica Scandinavica. 1997;68:511–514. - PubMed
-
- Frame G, Bretland KA, Dengler-Crish CM. Mechanistic complexities of bone loss in Alzheimer’s disease: a review. Connect. Tissue Res. 2020;61:4–18. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

