Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT)
- PMID: 34704059
- PMCID: PMC8496086
- DOI: 10.1039/d1cb00134e
Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT)
Abstract
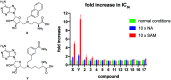
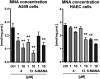
Nicotinamide N-methyltransferase (NNMT) methylates nicotinamide to form 1-methylnicotinamide (MNA) using S-adenosyl-l-methionine (SAM) as the methyl donor. The complexity of the role of NNMT in healthy and disease states is slowly being elucidated and provides an indication that NNMT may be an interesting therapeutic target for a variety of diseases including cancer, diabetes, and obesity. Most inhibitors of NNMT described to date are structurally related to one or both of its substrates. In the search for structurally diverse NNMT inhibitors, an mRNA display screening technique was used to identify macrocyclic peptides which bind to NNMT. Several of the cyclic peptides identified in this manner show potent inhibition of NNMT with IC50 values as low as 229 nM. The peptides were also found to downregulate MNA production in cellular assays. Interestingly, substrate competition experiments reveal that these cyclic peptide inhibitors are noncompetitive with either SAM or NA indicating they may be the first allosteric inhibitors reported for NNMT.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

Similar articles
-
Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics.J Med Chem. 2021 Sep 9;64(17):12938-12963. doi: 10.1021/acs.jmedchem.1c01094. Epub 2021 Aug 23. J Med Chem. 2021. PMID: 34424711 Free PMC article.
-
Selective and membrane-permeable small molecule inhibitors of nicotinamide N-methyltransferase reverse high fat diet-induced obesity in mice.Biochem Pharmacol. 2018 Jan;147:141-152. doi: 10.1016/j.bcp.2017.11.007. Epub 2017 Nov 15. Biochem Pharmacol. 2018. PMID: 29155147 Free PMC article.
-
A Rapid and Efficient Assay for the Characterization of Substrates and Inhibitors of Nicotinamide N-Methyltransferase.Biochemistry. 2016 Sep 20;55(37):5307-15. doi: 10.1021/acs.biochem.6b00733. Epub 2016 Sep 9. Biochemistry. 2016. PMID: 27570878
-
Complex roles of nicotinamide N-methyltransferase in cancer progression.Cell Death Dis. 2022 Mar 25;13(3):267. doi: 10.1038/s41419-022-04713-z. Cell Death Dis. 2022. PMID: 35338115 Free PMC article. Review.
-
Nicotinamide N-methyl transferase (NNMT): An emerging therapeutic target.Drug Discov Today. 2021 Nov;26(11):2699-2706. doi: 10.1016/j.drudis.2021.05.011. Epub 2021 May 21. Drug Discov Today. 2021. PMID: 34029690 Review.
Cited by
-
It Takes Two to Tango: The Interplay between Prostate Cancer and Its Microenvironment from an Epigenetic Perspective.Cancers (Basel). 2024 Jan 10;16(2):294. doi: 10.3390/cancers16020294. Cancers (Basel). 2024. PMID: 38254784 Free PMC article. Review.
-
Oncotherapeutic Strategies in Early Onset Colorectal Cancer.Cancers (Basel). 2023 Jan 16;15(2):552. doi: 10.3390/cancers15020552. Cancers (Basel). 2023. PMID: 36672501 Free PMC article. Review.
-
Proteomics Profiling of Bladder Cancer Tissues from Early to Advanced Stages Reveals NNMT and GALK1 as Biomarkers for Early Detection and Prognosis of BCa.Int J Mol Sci. 2023 Oct 6;24(19):14938. doi: 10.3390/ijms241914938. Int J Mol Sci. 2023. PMID: 37834386 Free PMC article.
-
NAD+ Homeostasis and NAD+-Consuming Enzymes: Implications for Vascular Health.Antioxidants (Basel). 2023 Feb 4;12(2):376. doi: 10.3390/antiox12020376. Antioxidants (Basel). 2023. PMID: 36829935 Free PMC article. Review.
-
Melanogenesis and the Targeted Therapy of Melanoma.Biomolecules. 2022 Dec 14;12(12):1874. doi: 10.3390/biom12121874. Biomolecules. 2022. PMID: 36551302 Free PMC article. Review.
References
-
- Alston T. A. Abeles R. H. Substrate specificity of nicotinamide methyltransferase isolated from porcine liver. Arch. Biochem. Biophys. 1988;260:601–608. - PubMed
-
- van Haren M. J. Sastre Toraño J. Sartini D. Emanuelli M. Parsons R. B. Martin N. I. A Rapid and Efficient Assay for the Characterization of Substrates and Inhibitors of Nicotinamide N-Methyltransferase. Biochemistry. 2016;55:5307–5315. - PubMed
-
- Eckert M. A. Coscia F. Chryplewicz A. Chang J. W. Hernandez K. M. Pan S. Tienda S. M. Nahotko D. A. Li G. Blaženović I. Lastra R. R. Curtis M. Yamada S. D. Perets R. McGregor S. M. Andrade J. Fiehn O. Moellering R. E. Mann M. Lengyel E. Proteomics reveals NNMT as a master metabolic regulator of cancer-associated fibroblasts. Nature. 2019;569:723–728. - PMC - PubMed
-
- Kanakkanthara A. Kurmi K. Ekstrom T. L. Hou X. Purfeerst E. R. Heinzen E. P. Correia C. Huntoon C. J. O’Brien D. Wahner Hendrickson A. E. Dowdy S. C. Li H. Oberg A. L. Hitosugi T. Kaufmann S. H. Weroha S. J. Karnitz L. M. BRCA1 Deficiency Upregulates NNMT, Which Reprograms Metabolism and Sensitizes Ovarian Cancer Cells to Mitochondrial Metabolic Targeting Agents. Cancer Res. 2019;79:5920–5929. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

