Insight into the Binding and Hydrolytic Preferences of hNudt16 Based on Nucleotide Diphosphate Substrates
- PMID: 34681586
- PMCID: PMC8535469
- DOI: 10.3390/ijms222010929
Insight into the Binding and Hydrolytic Preferences of hNudt16 Based on Nucleotide Diphosphate Substrates
Abstract
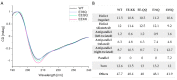
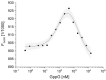
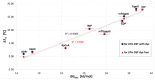
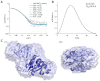
Nudt16 is a member of the NUDIX family of hydrolases that show specificity towards substrates consisting of a nucleoside diphosphate linked to another moiety X. Several substrates for hNudt16 and various possible biological functions have been reported. However, some of these reports contradict each other and studies comparing the substrate specificity of the hNudt16 protein are limited. Therefore, we quantitatively compared the affinity of hNudt16 towards a set of previously published substrates, as well as identified novel potential substrates. Here, we show that hNudt16 has the highest affinity towards IDP and GppG, with Kd below 100 nM. Other tested ligands exhibited a weaker affinity of several orders of magnitude. Among the investigated compounds, only IDP, GppG, m7GppG, AppA, dpCoA, and NADH were hydrolyzed by hNudt16 with a strong substrate preference for inosine or guanosine containing compounds. A new identified substrate for hNudt16, GppG, which binds the enzyme with an affinity comparable to that of IDP, suggests another potential regulatory role of this protein. Molecular docking of hNudt16-ligand binding inside the hNudt16 pocket revealed two binding modes for representative substrates. Nucleobase stabilization by Π stacking interactions with His24 has been associated with strong binding of hNudt16 substrates.
Keywords: DSF; GppG; IDP; MST; SAXS; dinucleoside diphosphates; hNudt16; nudix family.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Hydrolytic activity of human Nudt16 enzyme on dinucleotide cap analogs and short capped oligonucleotides.RNA. 2018 May;24(5):633-642. doi: 10.1261/rna.065698.118. Epub 2018 Feb 26. RNA. 2018. PMID: 29483298 Free PMC article.
-
hNUDT16: a universal decapping enzyme for small nucleolar RNA and cytoplasmic mRNA.Protein Cell. 2011 Jan;2(1):64-73. doi: 10.1007/s13238-011-1009-2. Epub 2011 Feb 20. Protein Cell. 2011. PMID: 21337011 Free PMC article.
-
Substrate specificity characterization for eight putative nudix hydrolases. Evaluation of criteria for substrate identification within the Nudix family.Proteins. 2016 Dec;84(12):1810-1822. doi: 10.1002/prot.25163. Epub 2016 Oct 1. Proteins. 2016. PMID: 27618147 Free PMC article.
-
Structures and mechanisms of Nudix hydrolases.Arch Biochem Biophys. 2005 Jan 1;433(1):129-43. doi: 10.1016/j.abb.2004.08.017. Arch Biochem Biophys. 2005. PMID: 15581572 Review.
-
The plant Nudix hydrolase family.Acta Biochim Pol. 2008;55(4):663-71. Epub 2008 Dec 16. Acta Biochim Pol. 2008. PMID: 19081844 Review.
Cited by
-
Application of Mammalian Nudix Enzymes to Capped RNA Analysis.Pharmaceuticals (Basel). 2024 Sep 11;17(9):1195. doi: 10.3390/ph17091195. Pharmaceuticals (Basel). 2024. PMID: 39338357 Free PMC article. Review.
-
Identification and validation of novel genes related to immune microenvironment in polycystic ovary syndrome.Medicine (Baltimore). 2024 Oct 25;103(43):e40229. doi: 10.1097/MD.0000000000040229. Medicine (Baltimore). 2024. PMID: 39470566 Free PMC article.
-
The Role of Hydrolases in Biology and Xenobiotics Metabolism.Int J Mol Sci. 2022 Apr 28;23(9):4870. doi: 10.3390/ijms23094870. Int J Mol Sci. 2022. PMID: 35563260 Free PMC article.
-
N2 modified cap analogues as translation inhibitors and substrates for preparation of therapeutic mRNA.Eur Biophys J. 2023 Oct;52(6-7):511-519. doi: 10.1007/s00249-023-01676-7. Epub 2023 Sep 1. Eur Biophys J. 2023. PMID: 37656232 Free PMC article. Review.
-
The potential of N2-modified cap analogues for precise genetic manipulation through mRNA engineering.Front Mol Biosci. 2024 Feb 6;10:1269028. doi: 10.3389/fmolb.2023.1269028. eCollection 2023. Front Mol Biosci. 2024. PMID: 38380271 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

