CellProfiler 4: improvements in speed, utility and usability
- PMID: 34507520
- PMCID: PMC8431850
- DOI: 10.1186/s12859-021-04344-9
CellProfiler 4: improvements in speed, utility and usability
Abstract
Background: Imaging data contains a substantial amount of information which can be difficult to evaluate by eye. With the expansion of high throughput microscopy methodologies producing increasingly large datasets, automated and objective analysis of the resulting images is essential to effectively extract biological information from this data. CellProfiler is a free, open source image analysis program which enables researchers to generate modular pipelines with which to process microscopy images into interpretable measurements.
Results: Herein we describe CellProfiler 4, a new version of this software with expanded functionality. Based on user feedback, we have made several user interface refinements to improve the usability of the software. We introduced new modules to expand the capabilities of the software. We also evaluated performance and made targeted optimizations to reduce the time and cost associated with running common large-scale analysis pipelines.
Conclusions: CellProfiler 4 provides significantly improved performance in complex workflows compared to previous versions. This release will ensure that researchers will have continued access to CellProfiler's powerful computational tools in the coming years.
Keywords: Bioimaging; Image analysis; Image quantitation; Image segmentation; Microscopy.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

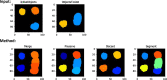
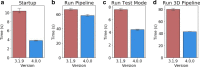
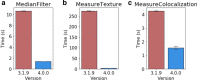


Similar articles
-
CellProfiler 3.0: Next-generation image processing for biology.PLoS Biol. 2018 Jul 3;16(7):e2005970. doi: 10.1371/journal.pbio.2005970. eCollection 2018 Jul. PLoS Biol. 2018. PMID: 29969450 Free PMC article.
-
ImageJ and CellProfiler: Complements in Open-Source Bioimage Analysis.Curr Protoc. 2021 May;1(5):e89. doi: 10.1002/cpz1.89. Curr Protoc. 2021. PMID: 34038030 Free PMC article.
-
easyXpress: An R package to analyze and visualize high-throughput C. elegans microscopy data generated using CellProfiler.PLoS One. 2021 Aug 12;16(8):e0252000. doi: 10.1371/journal.pone.0252000. eCollection 2021. PLoS One. 2021. PMID: 34383778 Free PMC article.
-
Workflows for microscopy image analysis and cellular phenotyping.J Biotechnol. 2017 Nov 10;261:70-75. doi: 10.1016/j.jbiotec.2017.07.019. Epub 2017 Jul 27. J Biotechnol. 2017. PMID: 28757289 Review.
-
Review of free software tools for image analysis of fluorescence cell micrographs.J Microsc. 2015 Jan;257(1):39-53. doi: 10.1111/jmi.12184. Epub 2014 Oct 31. J Microsc. 2015. PMID: 25359577 Review.
Cited by
-
Endosomal protein DENND10/FAM45A integrates extracellular vesicle release with cancer cell migration.BMC Biol. 2024 Jul 10;22(1):154. doi: 10.1186/s12915-024-01948-4. BMC Biol. 2024. PMID: 38987765 Free PMC article.
-
Differential intra-host infection kinetics in Aedes aegypti underlie superior transmissibility of African relative to Asian Zika virus.mSphere. 2023 Dec 20;8(6):e0054523. doi: 10.1128/msphere.00545-23. Epub 2023 Nov 9. mSphere. 2023. PMID: 37943061 Free PMC article.
-
Initial Characterization of WDR5B Reveals a Role in the Proliferation of Retinal Pigment Epithelial Cells.Cells. 2024 Jul 13;13(14):1189. doi: 10.3390/cells13141189. Cells. 2024. PMID: 39056772 Free PMC article.
-
High-throughput image processing software for the study of nuclear architecture and gene expression.Sci Rep. 2024 Aug 8;14(1):18426. doi: 10.1038/s41598-024-66600-1. Sci Rep. 2024. PMID: 39117696 Free PMC article.
-
Coxiella burnetii protein CBU2016 supports CCV expansion.Pathog Dis. 2024 Feb 7;82:ftae018. doi: 10.1093/femspd/ftae018. Pathog Dis. 2024. PMID: 39138067 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

