Circulating circRNA as biomarkers for dilated cardiomyopathy etiology
- PMID: 34498126
- PMCID: PMC8599237
- DOI: 10.1007/s00109-021-02119-6
Circulating circRNA as biomarkers for dilated cardiomyopathy etiology
Abstract
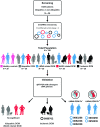
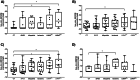
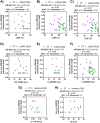
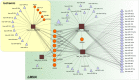
Dilated cardiomyopathy (DCM) is the third most common cause of heart failure. The multidisciplinary nature of testing - involving genetics, imaging, or cardiovascular techniques - makes its diagnosis challenging. Novel and reliable biomarkers are needed for early identification and tailored personalized management. Peripheral circular RNAs (circRNAs), a leading research topic, remain mostly unexplored in DCM. We aimed to assess whether peripheral circRNAs are expressed differentially among etiology-based DCM. The study was based on a case-control multicentric study. We enrolled 130 subjects: healthy controls (n = 20), idiopathic DCM (n = 30), ischemic DCM (n = 20), and familial DCM patients which included pathogen variants of (i) LMNA gene (n = 30) and (ii) BCL2-associated athanogene 3 (BAG3) gene (n = 30). Differentially expressed circRNAs were analyzed in plasma samples by quantitative RT-PCR and correlated to relevant systolic and diastolic parameters. The pathophysiological implications were explored through bioinformatics tools. Four circRNAs were overexpressed compared to controls: hsa_circ_0003258, hsa_circ_0051238, and hsa_circ_0051239 in LMNA-related DCM and hsa_circ_0089762 in the ischemic DCM cohort. The obtained areas under the curve confirm the discriminative capacity of circRNAs. The circRNAs correlated with some diastolic and systolic echocardiographic parameters with notable diagnostic potential in DCM. Circulating circRNAs may be helpful for the etiology-based diagnosis of DCM as a non-invasive biomarker. KEY MESSAGES: The limitations of cardiac diagnostic imaging and the absence of a robust biomarker reveal the need for a diagnostic tool for dilated cardiomyopathy (DCM). The circular RNA (circRNA) expression pattern is paramount for categorizing the DCM etiologies. Our peripheral circRNAs fingerprint discriminates between various among etiology-based DCM and correlates with some echocardiographic parameters. We provide a potential non-invasive biomarker for the etiology-based diagnosis of LMNA-related DCM and ischemic DCM.
Keywords: Circulating circular RNA; Ischemic-dilated cardiomyopathy; Lamin A/C-dilated cardiomyopathy.
© 2021. The Author(s).
Conflict of interest statement
We know of no conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome. The study protocol was approved by the Andalusian Biomedical Research Ethics committee. The study was performed in full compliance with the Declaration of Helsinki. Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients for paper publication.
Figures
Similar articles
-
Plasma microRNAs as biomarkers for Lamin A/C-related dilated cardiomyopathy.J Mol Med (Berl). 2018 Aug;96(8):845-856. doi: 10.1007/s00109-018-1666-1. Epub 2018 Jul 14. J Mol Med (Berl). 2018. PMID: 30008018
-
A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma.J Transl Med. 2019 Feb 18;17(1):50. doi: 10.1186/s12967-019-1800-z. J Transl Med. 2019. PMID: 30777071 Free PMC article.
-
Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children.J Clin Lab Anal. 2019 Nov;33(9):e22998. doi: 10.1002/jcla.22998. Epub 2019 Aug 20. J Clin Lab Anal. 2019. PMID: 31429492 Free PMC article.
-
Emerging role of microRNAs in dilated cardiomyopathy: evidence regarding etiology.Transl Res. 2020 Jan;215:86-101. doi: 10.1016/j.trsl.2019.08.007. Epub 2019 Aug 23. Transl Res. 2020. PMID: 31505160 Review.
-
Plain Language Summary of Publication of the safety and efficacy of ARRY-371797 in people with dilated cardiomyopathy and a faulty LMNA gene.Future Cardiol. 2023 Feb;19(2):55-63. doi: 10.2217/fca-2022-0099. Epub 2023 Jan 31. Future Cardiol. 2023. PMID: 36718638 Review.
Cited by
-
Transcriptome studies of inherited dilated cardiomyopathies.Mamm Genome. 2023 Jun;34(2):312-322. doi: 10.1007/s00335-023-09978-z. Epub 2023 Feb 7. Mamm Genome. 2023. PMID: 36749382 Free PMC article. Review.
-
Identification and verification of circRNA biomarkers for coronary artery disease based on WGCNA and the LASSO algorithm.BMC Cardiovasc Disord. 2024 Jun 17;24(1):305. doi: 10.1186/s12872-024-03972-2. BMC Cardiovasc Disord. 2024. PMID: 38880872 Free PMC article.
-
Diagnosis of acute myocardial infarction using a combination of circulating circular RNA cZNF292 and clinical information based on machine learning.MedComm (2020). 2023 Jun 13;4(3):e299. doi: 10.1002/mco2.299. eCollection 2023 Jun. MedComm (2020). 2023. PMID: 37323876 Free PMC article.
-
Exploring the Multifaceted Biologically Relevant Roles of circRNAs: From Regulation, Translation to Biomarkers.Cells. 2023 Dec 10;12(24):2813. doi: 10.3390/cells12242813. Cells. 2023. PMID: 38132133 Free PMC article. Review.
-
A Comprehensive Outlook on Dilated Cardiomyopathy (DCM): State-Of-The-Art Developments with Special Emphasis on OMICS-Based Approaches.J Cardiovasc Dev Dis. 2022 Jun 1;9(6):174. doi: 10.3390/jcdd9060174. J Cardiovasc Dev Dis. 2022. PMID: 35735803 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

