Prognostic correlations with the microbiome of breast cancer subtypes
- PMID: 34482363
- PMCID: PMC8418604
- DOI: 10.1038/s41419-021-04092-x
Prognostic correlations with the microbiome of breast cancer subtypes
Abstract
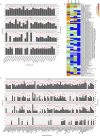
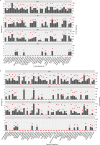
Alterations to the natural microbiome are linked to different diseases, and the presence or absence of specific microbes is directly related to disease outcomes. We performed a comprehensive analysis with unique cohorts of the four subtypes of breast cancer (BC) characterized by their microbial signatures, using a pan-pathogen microarray strategy. The signature (includes viruses, bacteria, fungi, and parasites) of each tumor subtype was correlated with clinical data to identify microbes with prognostic potential. The subtypes of BC had specific viromes and microbiomes, with ER+ and TN tumors showing the most and least diverse microbiome, respectively. The specific microbial signatures allowed discrimination between different BC subtypes. Furthermore, we demonstrated correlations between the presence and absence of specific microbes in BC subtypes with the clinical outcomes. This study provides a comprehensive map of the oncobiome of BC subtypes, with insights into disease prognosis that can be critical for precision therapeutic intervention strategies.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Association of molecular subtypes with breast cancer risk factors: a case-only analysis.Eur J Cancer Prev. 2015 Nov;24(6):484-90. doi: 10.1097/CEJ.0000000000000111. Eur J Cancer Prev. 2015. PMID: 25494290
-
Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers.Breast Cancer Res. 2014 Dec 31;16(6):3416. doi: 10.1186/s13058-014-0492-9. Breast Cancer Res. 2014. PMID: 25919761 Free PMC article.
-
Breast cancer molecular subtypes and survival in a hospital-based sample in Puerto Rico.Cancer Med. 2013 Jun;2(3):343-50. doi: 10.1002/cam4.78. Epub 2013 Apr 18. Cancer Med. 2013. PMID: 23930211 Free PMC article.
-
Impact of molecular subtypes classification concordance between preoperative core needle biopsy and surgical specimen on early breast cancer management: Single-institution experience and review of published literature.Eur J Surg Oncol. 2017 Apr;43(4):642-648. doi: 10.1016/j.ejso.2016.10.025. Epub 2016 Nov 17. Eur J Surg Oncol. 2017. PMID: 27889196 Review.
-
Potential prognostic tumor biomarkers in triple-negative breast carcinoma.Beijing Da Xue Xue Bao Yi Xue Ban. 2012 Oct 18;44(5):666-72. Beijing Da Xue Xue Bao Yi Xue Ban. 2012. PMID: 23073572 Review.
Cited by
-
The Influence of Microbiota on Breast Cancer: A Review.Cancers (Basel). 2024 Oct 13;16(20):3468. doi: 10.3390/cancers16203468. Cancers (Basel). 2024. PMID: 39456562 Free PMC article. Review.
-
Cytokine levels in breast cancer are highly dependent on cytomegalovirus (CMV) status.Breast Cancer Res Treat. 2024 Dec;208(3):631-641. doi: 10.1007/s10549-024-07459-8. Epub 2024 Aug 22. Breast Cancer Res Treat. 2024. PMID: 39172306 Free PMC article.
-
Guideline for designing microbiome studies in neoplastic diseases.Geroscience. 2024 Oct;46(5):4037-4057. doi: 10.1007/s11357-024-01255-4. Epub 2024 Jun 26. Geroscience. 2024. PMID: 38922379 Free PMC article. Review.
-
Residents or Tourists: Is the Lactating Mammary Gland Colonized by Residential Microbiota?Microorganisms. 2024 May 17;12(5):1009. doi: 10.3390/microorganisms12051009. Microorganisms. 2024. PMID: 38792838 Free PMC article. Review.
-
Promising dawn in tumor microenvironment therapy: engineering oral bacteria.Int J Oral Sci. 2024 Mar 13;16(1):24. doi: 10.1038/s41368-024-00282-3. Int J Oral Sci. 2024. PMID: 38472176 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

