Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance
- PMID: 34412018
- PMCID: PMC8341816
- DOI: 10.1016/j.watres.2021.117516
Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance
Abstract
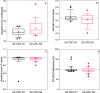
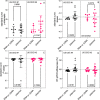
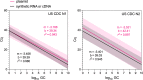
Due to the coronavirus disease 2019 (COVID-19) pandemic, wastewater surveillance has become an important tool for monitoring the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) within communities. In particular, reverse transcription-quantitative PCR (RT-qPCR) has been used to generate large datasets aimed at detecting and quantifying SARS-CoV-2 RNA in wastewater. Although RT-qPCR is rapid and sensitive, there is no standard method yet, there are no certified quantification standards, and experiments are conducted using different assays, reagents, instruments, and data analysis protocols. These variations can induce errors in quantitative data reports, thereby potentially misleading interpretations, and conclusions. We review the SARS-CoV-2 wastewater surveillance literature focusing on variability of RT-qPCR data as revealed by inconsistent standard curves and associated parameters. We find that variation in these parameters and deviations from best practices, as described in the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines suggest a frequent lack of reproducibility and reliability in quantitative measurements of SARS-CoV-2 RNA in wastewater.
Keywords: RT-qPCR; assay validity; quality assurance; quality control; standard curve; wastewater surveillance.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Comparison of Different Reverse Transcriptase-Polymerase Chain Reaction-Based Methods for Wastewater Surveillance of SARS-CoV-2: Exploratory Study.JMIR Public Health Surveill. 2024 Aug 19;10:e53175. doi: 10.2196/53175. JMIR Public Health Surveill. 2024. PMID: 39158943 Free PMC article.
-
The Inhibition and Variability of Two Different RT-qPCR Assays Used for Quantifying SARS-CoV-2 RNA in Wastewater.Food Environ Virol. 2023 Mar;15(1):71-81. doi: 10.1007/s12560-022-09542-z. Epub 2023 Feb 15. Food Environ Virol. 2023. PMID: 36790663 Free PMC article.
-
RT-qPCR and ATOPlex sequencing for the sensitive detection of SARS-CoV-2 RNA for wastewater surveillance.Water Res. 2022 Jul 15;220:118621. doi: 10.1016/j.watres.2022.118621. Epub 2022 May 16. Water Res. 2022. PMID: 35665675 Free PMC article.
-
Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance.Sci Total Environ. 2022 Jan 20;805:149877. doi: 10.1016/j.scitotenv.2021.149877. Epub 2021 Aug 25. Sci Total Environ. 2022. PMID: 34818780 Free PMC article. Review.
-
A review on detection of SARS-CoV-2 RNA in wastewater in light of the current knowledge of treatment process for removal of viral fragments.J Environ Manage. 2021 Dec 1;299:113563. doi: 10.1016/j.jenvman.2021.113563. Epub 2021 Aug 19. J Environ Manage. 2021. PMID: 34488114 Free PMC article. Review.
Cited by
-
PCR standard curve quantification in an extensive wastewater surveillance program: results from the Dutch SARS-CoV-2 wastewater surveillance.Front Public Health. 2023 Nov 2;11:1141494. doi: 10.3389/fpubh.2023.1141494. eCollection 2023. Front Public Health. 2023. PMID: 38026384 Free PMC article.
-
Digital PCR can augment the interpretation of RT-qPCR Cq values for SARS-CoV-2 diagnostics.Methods. 2022 May;201:5-14. doi: 10.1016/j.ymeth.2021.08.006. Epub 2021 Aug 26. Methods. 2022. PMID: 34454016 Free PMC article.
-
Wastewater surveillance in smaller college communities may aid future public health initiatives.PLoS One. 2022 Sep 16;17(9):e0270385. doi: 10.1371/journal.pone.0270385. eCollection 2022. PLoS One. 2022. PMID: 36112629 Free PMC article.
-
Occurrence and Genetic Characterization of Grapevine Pinot Gris Virus in Russia.Plants (Basel). 2022 Apr 13;11(8):1061. doi: 10.3390/plants11081061. Plants (Basel). 2022. PMID: 35448789 Free PMC article.
-
Comparison of Reverse Transcription (RT)-Quantitative PCR and RT-Droplet Digital PCR for Detection of Genomic and Subgenomic SARS-CoV-2 RNA.Microbiol Spectr. 2023 Mar 21;11(2):e0415922. doi: 10.1128/spectrum.04159-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36943067 Free PMC article.
References
-
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021;761 - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: Methods optimization and quality control are crucial for generating reliable public health information. Curr Opin Environ Sci Health. 2020;17:82–93. - PMC - PubMed
-
- Ahmed W., Simpson S., Bertsch P., Bibby K., Bivins A., Blackall L., Bofill-Mas S., Bosch A., Brandao J., Choi P., Ciesielski M., Donner E., D'Souza N., Farnleitner A., Gerrity D., Gonzalez R., Griffith J., Gyawali P., Haas C., Hamilton K., Hapuarachchi C., Harwood V., Haque R., Jackson G., Khan S., Khan W., Kitajima M., Korajkic A., La Rosa G., Layton B., Lipp E., McLellan S., McMinn B., Medema G., Metcalfe S., Meijer W., Mueller J., Murphy H., Naughton C., Noble R., Payyappat S., Petterson S., Pitkanen T., Rajal V., Reyneke B., Roman F., Rose J., Rusinol M., Sadowsky M., Sala-Comorera L., Setoh Y.X., Sherchan S., Sirikanchana K., Smith W., Steele J., Sabburg R., Symonds E., Thai P., Thomas K., Tynan J., Toze S., Thompson J., Whiteley A., Wong J., Sano D., Wuertz S., Xagoraraki I., Zhang Q., Zimmer-Faust A., Shanks O. Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Preprints. 2021 doi: 10.20944/preprints202104.0481.v1. 2021040481. - DOI - PMC - PubMed
-
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Gurol Z.C., Chakraborty C., Costa F., Curcio S., de los reyes F.L., III, Delgado Vela J., Farkas K., Fernazdez-Casi X., Gerba C., Gerrity D., Girones R., Gonzzalez R., Haramoto E., Harris A., Holden P.A., Ispam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., Mclellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Noble R.T., van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., van der Voorn T., Wiggington K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

