PHROG: families of prokaryotic virus proteins clustered using remote homology
- PMID: 34377978
- PMCID: PMC8341000
- DOI: 10.1093/nargab/lqab067
PHROG: families of prokaryotic virus proteins clustered using remote homology
Abstract
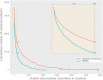
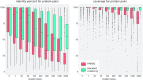
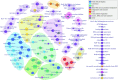
Viruses are abundant, diverse and ancestral biological entities. Their diversity is high, both in terms of the number of different protein families encountered and in the sequence heterogeneity of each protein family. The recent increase in sequenced viral genomes constitutes a great opportunity to gain new insights into this diversity and consequently urges the development of annotation resources to help functional and comparative analysis. Here, we introduce PHROG (Prokaryotic Virus Remote Homologous Groups), a library of viral protein families generated using a new clustering approach based on remote homology detection by HMM profile-profile comparisons. Considering 17 473 reference (pro)viruses of prokaryotes, 868 340 of the total 938 864 proteins were grouped into 38 880 clusters that proved to be a 2-fold deeper clustering than using a classical strategy based on BLAST-like similarity searches, and yet to remain homogeneous. Manual inspection of similarities to various reference sequence databases led to the annotation of 5108 clusters (containing 50.6 % of the total protein dataset) with 705 different annotation terms, included in 9 functional categories, specifically designed for viruses. Hopefully, PHROG will be a useful tool to better annotate future prokaryotic viral sequences thus helping the scientific community to better understand the evolution and ecology of these entities.
© The Author(s) 2021. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


Similar articles
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
VOGDB-Database of Virus Orthologous Groups.Viruses. 2024 Jul 25;16(8):1191. doi: 10.3390/v16081191. Viruses. 2024. PMID: 39205165 Free PMC article.
-
Prokaryotic Virus Orthologous Groups (pVOGs): a resource for comparative genomics and protein family annotation.Nucleic Acids Res. 2017 Jan 4;45(D1):D491-D498. doi: 10.1093/nar/gkw975. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789703 Free PMC article.
-
Filling-in void and sparse regions in protein sequence space by protein-like artificial sequences enables remarkable enhancement in remote homology detection capability.J Mol Biol. 2014 Feb 20;426(4):962-79. doi: 10.1016/j.jmb.2013.11.026. Epub 2013 Dec 4. J Mol Biol. 2014. PMID: 24316367
-
Large-Scale Sequencing: The Future of Genomic Sciences? This report is based on a colloquium, sponsored by the American Academy of Microbiology, convened September 2008 in Washington, DC.Washington (DC): American Society for Microbiology; 2009. Washington (DC): American Society for Microbiology; 2009. PMID: 33119235 Free Books & Documents. Review.
Cited by
-
Analysis of viromes and microbiomes from pig fecal samples reveals that phages and prophages rarely carry antibiotic resistance genes.ISME Commun. 2021 Oct 25;1(1):55. doi: 10.1038/s43705-021-00054-8. ISME Commun. 2021. PMID: 37938642 Free PMC article.
-
Viromes vs. mixed community metagenomes: choice of method dictates interpretation of viral community ecology.Microbiome. 2024 Oct 7;12(1):195. doi: 10.1186/s40168-024-01905-x. Microbiome. 2024. PMID: 39375774 Free PMC article.
-
Identification of Huge Phages from Wastewater Metagenomes.Viruses. 2023 Nov 28;15(12):2330. doi: 10.3390/v15122330. Viruses. 2023. PMID: 38140571 Free PMC article.
-
Deciphering Phage-Host Specificity Based on the Association of Phage Depolymerases and Bacterial Surface Glycan with Deep Learning.bioRxiv [Preprint]. 2023 Jun 16:2023.06.16.545366. doi: 10.1101/2023.06.16.545366. bioRxiv. 2023. PMID: 37503040 Free PMC article. Preprint.
-
Isolation and genomic characterization of Staphylococcus aureus bacteriophages from Chennai, India.Microbiol Resour Announc. 2024 Apr 11;13(4):e0120923. doi: 10.1128/mra.01209-23. Epub 2024 Mar 8. Microbiol Resour Announc. 2024. PMID: 38456698 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

