Insights into the Binding of Intrinsically Disordered Proteins from Molecular Dynamics Simulation
- PMID: 34354764
- PMCID: PMC8336759
- DOI: 10.1002/wcms.1167
Insights into the Binding of Intrinsically Disordered Proteins from Molecular Dynamics Simulation
Abstract
Intrinsically disordered proteins (IDPs) are a class of protein that, in the native state, possess no well-defined secondary or tertiary structure, existing instead as dynamic ensembles of conformations. They are biologically important, with approximately 20% of all eukaryotic proteins disordered, and found at the heart of many biochemical networks. To fulfil their biological roles, many IDPs need to bind to proteins and/or nucleic acids. And while unstructured in solution, IDPs typically fold into a well-defined three-dimensional structure upon interaction with a binding partner. The flexibility and structural diversity inherent to IDPs makes this coupled folding and binding difficult to study at atomic resolution by experiment alone, and computer simulation currently offers perhaps the best opportunity to understand this process. But simulation of coupled folding and binding is itself extremely challenging; these molecules are large and highly flexible, and their binding partners, such as DNA or cyclins, are also often large. Therefore, their study requires either or both simplified representations and advanced enhanced sampling schemes. It is not always clear that existing simulation techniques, optimized for studying folded proteins, are well-suited to IDPs. In this article, we examine the progress that has been made in the study of coupled folding and binding using molecular dynamics simulation. We summarise what has been learnt, and examine the state of the art in terms of both methodologies and models. We also consider the lessons to be learnt from advances in other areas of simulation and highlight the issues that remain of be addressed.
Figures
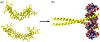
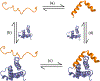

Similar articles
-
Complete Coupled Binding-Folding Pathway of the Intrinsically Disordered Transcription Factor Protein Brinker Revealed by Molecular Dynamics Simulations and Markov State Modeling.Biochemistry. 2018 Jul 31;57(30):4404-4420. doi: 10.1021/acs.biochem.8b00441. Epub 2018 Jul 19. Biochemistry. 2018. PMID: 29990433
-
Recent advances in atomic molecular dynamics simulation of intrinsically disordered proteins.Phys Chem Chem Phys. 2021 Jan 21;23(2):777-784. doi: 10.1039/d0cp05818a. Phys Chem Chem Phys. 2021. PMID: 33355572 Review.
-
Mechanism of Coupled Folding-upon-Binding of an Intrinsically Disordered Protein.J Am Chem Soc. 2020 Jun 24;142(25):11092-11101. doi: 10.1021/jacs.0c03217. Epub 2020 Jun 16. J Am Chem Soc. 2020. PMID: 32323533
-
Probing the dynamics of disorder.Prog Biophys Mol Biol. 2017 Sep;128:57-62. doi: 10.1016/j.pbiomolbio.2017.05.008. Epub 2017 May 26. Prog Biophys Mol Biol. 2017. PMID: 28554553 Review.
-
Hierarchical Ensembles of Intrinsically Disordered Proteins at Atomic Resolution in Molecular Dynamics Simulations.J Chem Theory Comput. 2020 Jan 14;16(1):725-737. doi: 10.1021/acs.jctc.9b00809. Epub 2019 Dec 26. J Chem Theory Comput. 2020. PMID: 31809054
Cited by
-
An energy decomposition and extrapolation scheme for evaluating electron transfer rate constants: a case study on electron self-exchange reactions of transition metal complexes.RSC Adv. 2023 Nov 1;13(46):32097-32103. doi: 10.1039/d3ra05784d. eCollection 2023 Oct 31. RSC Adv. 2023. PMID: 37920761 Free PMC article.
-
Discriminating binding mechanisms of an intrinsically disordered protein via a multi-state coarse-grained model.J Chem Phys. 2014 May 7;140(17):175102. doi: 10.1063/1.4873710. J Chem Phys. 2014. PMID: 24811666 Free PMC article.
-
III-defined concepts in chemistry: rigid force constants vs. compliance constants as bond strength descriptors for the triple bond in diboryne.Chem Sci. 2015 Jul 1;6(7):4086-4088. doi: 10.1039/c5sc01322d. Epub 2015 May 4. Chem Sci. 2015. PMID: 29218174 Free PMC article.
-
Protein disorder-to-order transition enhances the nucleosome-binding affinity of H1.Nucleic Acids Res. 2020 Jun 4;48(10):5318-5331. doi: 10.1093/nar/gkaa285. Nucleic Acids Res. 2020. PMID: 32356891 Free PMC article.
-
Full structural ensembles of intrinsically disordered proteins from unbiased molecular dynamics simulations.Commun Biol. 2021 Feb 23;4(1):243. doi: 10.1038/s42003-021-01759-1. Commun Biol. 2021. PMID: 33623120 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
