Investigating crosstalk between H3K27 acetylation and H3K4 trimethylation in CRISPR/dCas-based epigenome editing and gene activation
- PMID: 34354157
- PMCID: PMC8342468
- DOI: 10.1038/s41598-021-95398-5
Investigating crosstalk between H3K27 acetylation and H3K4 trimethylation in CRISPR/dCas-based epigenome editing and gene activation
Abstract
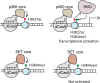
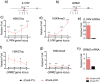
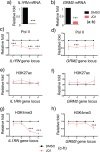
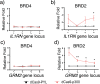
Epigenome editing methods enable the precise manipulation of epigenetic modifications, such as histone posttranscriptional modifications (PTMs), for uncovering their biological functions. While histone PTMs have been correlated with certain gene expression status, the causalities remain elusive. Histone H3 Lysine 27 acetylation (H3K27ac) and histone H3 Lysine 4 trimethylation (H3K4me3) are both associated with active genes, and located at active promoters and enhancers or around transcriptional start sites (TSSs). Although crosstalk between histone lysine acetylation and H3K4me3 has been reported, relationships between specific epigenetic marks during transcriptional activation remain largely unclear. Here, using clustered regularly interspaced short palindromic repeats (CRISPR)/dCas-based epigenome editing methods, we discovered that the ectopic introduction of H3K27ac in the promoter region lead to H3K4me3 enrichment around TSS and transcriptional activation, while H3K4me3 installation at the promoter cannot induce H3K27ac increase and failed to activate gene expression. Blocking the reading of H3K27ac by BRD proteins using inhibitor JQ1 abolished H3K27ac-induced H3K4me3 installation and downstream gene activation. Furthermore, we uncovered that BRD2, not BRD4, mediated H3K4me3 installation and gene activation upon H3K27ac writing. Our studies revealed the relationships between H3K27ac and H3K4me3 in gene activation process and demonstrated the application of CRISPR/dCas-based epigenome editing methods in elucidating the crosstalk between epigenetic mechanisms.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Histone modifications for human epigenome analysis.J Hum Genet. 2013 Jul;58(7):439-45. doi: 10.1038/jhg.2013.66. Epub 2013 Jun 6. J Hum Genet. 2013. PMID: 23739122 Review.
-
CRISPR/dCas9-mediated epigenetic modification reveals differential regulation of histone acetylation on Aspergillus niger secondary metabolite.Microbiol Res. 2021 Apr;245:126694. doi: 10.1016/j.micres.2020.126694. Epub 2021 Jan 6. Microbiol Res. 2021. PMID: 33482403
-
Lysine 27 of replication-independent histone H3.3 is required for Polycomb target gene silencing but not for gene activation.PLoS Genet. 2019 Jan 30;15(1):e1007932. doi: 10.1371/journal.pgen.1007932. eCollection 2019 Jan. PLoS Genet. 2019. PMID: 30699116 Free PMC article.
-
DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism.BMC Genomics. 2017 Dec 12;18(1):964. doi: 10.1186/s12864-017-4353-7. BMC Genomics. 2017. PMID: 29233090 Free PMC article.
-
Advances of epigenetic editing.Curr Opin Chem Biol. 2020 Aug;57:75-81. doi: 10.1016/j.cbpa.2020.04.020. Epub 2020 Jun 30. Curr Opin Chem Biol. 2020. PMID: 32619853 Review.
Cited by
-
Epigenetic control of pancreatic cancer metastasis.Cancer Metastasis Rev. 2023 Dec;42(4):1113-1131. doi: 10.1007/s10555-023-10132-z. Epub 2023 Sep 2. Cancer Metastasis Rev. 2023. PMID: 37659057 Free PMC article. Review.
-
Predicting the effect of CRISPR-Cas9-based epigenome editing.bioRxiv [Preprint]. 2024 Oct 16:2023.10.03.560674. doi: 10.1101/2023.10.03.560674. bioRxiv. 2024. PMID: 37873127 Free PMC article. Preprint.
-
Chromatin profiling and state predictions reveal insights into epigenetic regulation during early porcine development.Epigenetics Chromatin. 2024 May 21;17(1):16. doi: 10.1186/s13072-024-00542-w. Epigenetics Chromatin. 2024. PMID: 38773546 Free PMC article.
-
PLK-1 Interacting Checkpoint Helicase, PICH, Mediates Cellular Oxidative Stress Response.Epigenomes. 2022 Oct 18;6(4):36. doi: 10.3390/epigenomes6040036. Epigenomes. 2022. PMID: 36278682 Free PMC article.
-
Innate Immune Memory in Macrophages.Newborn (Clarksville). 2023 Jan-Mar;2(1):60-79. doi: 10.5005/jp-journals-11002-0058. Epub 2023 Apr 6. Newborn (Clarksville). 2023. PMID: 37206580 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

