The human cytomegalovirus protein pUL13 targets mitochondrial cristae architecture to increase cellular respiration during infection
- PMID: 34344827
- PMCID: PMC8364163
- DOI: 10.1073/pnas.2101675118
The human cytomegalovirus protein pUL13 targets mitochondrial cristae architecture to increase cellular respiration during infection
Abstract
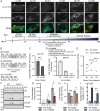
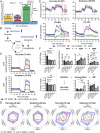
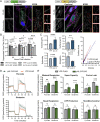
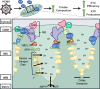
Viruses modulate mitochondrial processes during infection to increase biosynthetic precursors and energy output, fueling virus replication. In a surprising fashion, although it triggers mitochondrial fragmentation, the prevalent pathogen human cytomegalovirus (HCMV) increases mitochondrial metabolism through a yet-unknown mechanism. Here, we integrate molecular virology, metabolic assays, quantitative proteomics, and superresolution confocal microscopy to define this mechanism. We establish that the previously uncharacterized viral protein pUL13 is required for productive HCMV replication, targets the mitochondria, and functions to increase oxidative phosphorylation during infection. We demonstrate that pUL13 forms temporally tuned interactions with the mitochondrial contact site and cristae organizing system (MICOS) complex, a critical regulator of cristae architecture and electron transport chain (ETC) function. Stimulated emission depletion superresolution microscopy shows that expression of pUL13 alters cristae architecture. Indeed, using live-cell Seahorse assays, we establish that pUL13 alone is sufficient to increase cellular respiration, not requiring the presence of other viral proteins. Our findings address the outstanding question of how HCMV targets mitochondria to increase bioenergetic output and expands the knowledge of the intricate connection between mitochondrial architecture and ETC function.
Keywords: HCMV; metabolism; mitochondria; pUL13; proteomics.
Conflict of interest statement
The authors declare no competing interest.
Figures


Similar articles
-
HCMV strain- and cell type-specific alterations in membrane contact sites point to the convergent regulation of organelle remodeling.J Virol. 2024 Nov 19;98(11):e0109924. doi: 10.1128/jvi.01099-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480111
-
Nitric Oxide Circumvents Virus-Mediated Metabolic Regulation during Human Cytomegalovirus Infection.mBio. 2020 Dec 15;11(6):e02630-20. doi: 10.1128/mBio.02630-20. mBio. 2020. PMID: 33323506 Free PMC article.
-
Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection.J Virol. 2020 Jan 6;94(2):e01183-19. doi: 10.1128/JVI.01183-19. Print 2020 Jan 6. J Virol. 2020. PMID: 31694945 Free PMC article.
-
Access of viral proteins to mitochondria via mitochondria-associated membranes.Rev Med Virol. 2009 May;19(3):147-64. doi: 10.1002/rmv.611. Rev Med Virol. 2009. PMID: 19367604 Free PMC article. Review.
-
Prevention of cellular suicide by cytomegaloviruses.Viruses. 2012 Oct 2;4(10):1928-49. doi: 10.3390/v4101928. Viruses. 2012. PMID: 23202447 Free PMC article. Review.
Cited by
-
Role of immunometabolism during congenital cytomegalovirus infection.Immunometabolism (Cobham). 2023 Nov 28;5(4):e00034. doi: 10.1097/IN9.0000000000000034. eCollection 2023 Oct. Immunometabolism (Cobham). 2023. PMID: 38037590 Free PMC article.
-
Systems Biology of Virus-Host Protein Interactions: From Hypothesis Generation to Mechanisms of Replication and Pathogenesis.Annu Rev Virol. 2022 Sep 29;9(1):397-415. doi: 10.1146/annurev-virology-100520-011851. Epub 2022 May 16. Annu Rev Virol. 2022. PMID: 35576593 Free PMC article. Review.
-
The roles of autophagy and mitophagy in corneal pathology: current knowledge and future perspectives.Front Med (Lausanne). 2023 Apr 21;10:1064938. doi: 10.3389/fmed.2023.1064938. eCollection 2023. Front Med (Lausanne). 2023. PMID: 37153108 Free PMC article. Review.
-
HCMV strain- and cell type-specific alterations in membrane contact sites point to the convergent regulation of organelle remodeling.J Virol. 2024 Nov 19;98(11):e0109924. doi: 10.1128/jvi.01099-24. Epub 2024 Oct 31. J Virol. 2024. PMID: 39480111
-
Insights into the Role of VPS39 and Its Interaction with CP204L and A137R in ASFV Infection.Viruses. 2024 Sep 17;16(9):1478. doi: 10.3390/v16091478. Viruses. 2024. PMID: 39339953 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

