Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics
- PMID: 34301936
- PMCID: PMC8302612
- DOI: 10.1038/s41467-021-24461-6
Quantitative imaging of transcription in living Drosophila embryos reveals the impact of core promoter motifs on promoter state dynamics
Abstract
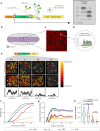
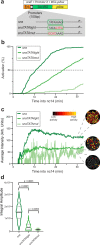
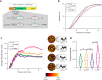
Genes are expressed in stochastic transcriptional bursts linked to alternating active and inactive promoter states. A major challenge in transcription is understanding how promoter composition dictates bursting, particularly in multicellular organisms. We investigate two key Drosophila developmental promoter motifs, the TATA box (TATA) and the Initiator (INR). Using live imaging in Drosophila embryos and new computational methods, we demonstrate that bursting occurs on multiple timescales ranging from seconds to minutes. TATA-containing promoters and INR-containing promoters exhibit distinct dynamics, with one or two separate rate-limiting steps respectively. A TATA box is associated with long active states, high rates of polymerase initiation, and short-lived, infrequent inactive states. In contrast, the INR motif leads to two inactive states, one of which relates to promoter-proximal polymerase pausing. Surprisingly, the model suggests pausing is not obligatory, but occurs stochastically for a subset of polymerases. Overall, our results provide a rationale for promoter switching during zygotic genome activation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Classification of Promoters Based on the Combination of Core Promoter Elements Exhibits Different Histone Modification Patterns.PLoS One. 2016 Mar 22;11(3):e0151917. doi: 10.1371/journal.pone.0151917. eCollection 2016. PLoS One. 2016. PMID: 27003446 Free PMC article.
-
Large distances separate coregulated genes in living Drosophila embryos.Proc Natl Acad Sci U S A. 2019 Jul 23;116(30):15062-15067. doi: 10.1073/pnas.1908962116. Epub 2019 Jul 8. Proc Natl Acad Sci U S A. 2019. PMID: 31285341 Free PMC article.
-
Computational identification and experimental characterization of preferred downstream positions in human core promoters.PLoS Comput Biol. 2021 Aug 12;17(8):e1009256. doi: 10.1371/journal.pcbi.1009256. eCollection 2021 Aug. PLoS Comput Biol. 2021. PMID: 34383743 Free PMC article.
-
The RNA polymerase II core promoter - the gateway to transcription.Curr Opin Cell Biol. 2008 Jun;20(3):253-9. doi: 10.1016/j.ceb.2008.03.003. Epub 2008 Apr 22. Curr Opin Cell Biol. 2008. PMID: 18436437 Free PMC article. Review.
-
Sample Preparation and Mounting of Drosophila Embryos for Multiview Light Sheet Microscopy.Methods Mol Biol. 2016;1478:189-202. doi: 10.1007/978-1-4939-6371-3_10. Methods Mol Biol. 2016. PMID: 27730582 Review.
Cited by
-
Scalable inference of transcriptional kinetic parameters from MS2 time series data.Bioinformatics. 2022 Jan 27;38(4):1030-1036. doi: 10.1093/bioinformatics/btab765. Bioinformatics. 2022. PMID: 34788793 Free PMC article.
-
Transcriptional bursting dynamics in gene expression.Front Genet. 2024 Sep 13;15:1451461. doi: 10.3389/fgene.2024.1451461. eCollection 2024. Front Genet. 2024. PMID: 39346775 Free PMC article. Review.
-
The miR-430 locus with extreme promoter density forms a transcription body during the minor wave of zygotic genome activation.Dev Cell. 2023 Jan 23;58(2):155-170.e8. doi: 10.1016/j.devcel.2022.12.007. Dev Cell. 2023. PMID: 36693321 Free PMC article.
-
Stochastic pausing at latent HIV-1 promoters generates transcriptional bursting.Nat Commun. 2021 Jul 23;12(1):4503. doi: 10.1038/s41467-021-24462-5. Nat Commun. 2021. PMID: 34301927 Free PMC article.
-
The method in the madness: Transcriptional control from stochastic action at the single-molecule scale.Curr Opin Struct Biol. 2024 Aug;87:102873. doi: 10.1016/j.sbi.2024.102873. Epub 2024 Jul 1. Curr Opin Struct Biol. 2024. PMID: 38954990 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

