Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture
- PMID: 34297726
- PMCID: PMC8336837
- DOI: 10.1371/journal.pbio.3001344
Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture
Abstract
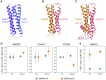
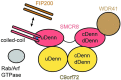
A major cause of familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) spectrum disorder is the hexanucleotide G4C2 repeat expansion in the first intron of the C9orf72 gene. Many underlying mechanisms lead to manifestation of disease that include toxic gain-of-function by repeat G4C2 RNAs, dipeptide repeat proteins, and a reduction of the C9orf72 gene product. The C9orf72 protein interacts with SMCR8 and WDR41 to form a trimeric complex and regulates multiple cellular pathways including autophagy. Here, we report the structure of the C9orf72-SMCR8 complex at 3.8 Å resolution using single-particle cryo-electron microscopy (cryo-EM). The structure reveals 2 distinct dimerization interfaces between C9orf72 and SMCR8 that involves an extensive network of interactions. Homology between C9orf72-SMCR8 and Folliculin-Folliculin Interacting Protein 2 (FLCN-FNIP2), a GTPase activating protein (GAP) complex, enabled identification of a key residue within the active site of SMCR8. Further structural analysis suggested that a coiled-coil region within the uDenn domain of SMCR8 could act as an interaction platform for other coiled-coil proteins, and its deletion reduced the interaction of the C9orf72-SMCR8 complex with FIP200 upon starvation. In summary, this study contributes toward our understanding of the biological function of the C9orf72-SMCR8 complex.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Cryo-EM structure of C9ORF72-SMCR8-WDR41 reveals the role as a GAP for Rab8a and Rab11a.Proc Natl Acad Sci U S A. 2020 May 5;117(18):9876-9883. doi: 10.1073/pnas.2002110117. Epub 2020 Apr 17. Proc Natl Acad Sci U S A. 2020. PMID: 32303654 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
The C9orf72-SMCR8-WDR41 complex is a GAP for small GTPases.Autophagy. 2020 Aug;16(8):1542-1543. doi: 10.1080/15548627.2020.1779473. Epub 2020 Jun 17. Autophagy. 2020. PMID: 32521185 Free PMC article.
-
Structure of the C9orf72 ARF GAP complex that is haploinsufficient in ALS and FTD.Nature. 2020 Sep;585(7824):251-255. doi: 10.1038/s41586-020-2633-x. Epub 2020 Aug 26. Nature. 2020. PMID: 32848248 Free PMC article.
-
The progress in C9orf72 research: ALS/FTD pathogenesis, functions and structure.Small GTPases. 2022 Jan;13(1):56-76. doi: 10.1080/21541248.2021.1892443. Epub 2021 Mar 5. Small GTPases. 2022. PMID: 33663328 Free PMC article. Review.
Cited by
-
Exploring dysregulated miRNAs in ALS: implications for disease pathogenesis and early diagnosis.Neurol Sci. 2024 Nov 21. doi: 10.1007/s10072-024-07840-x. Online ahead of print. Neurol Sci. 2024. PMID: 39570437
-
Structures and Functions of the Human GATOR1 Complex.Subcell Biochem. 2024;104:269-294. doi: 10.1007/978-3-031-58843-3_12. Subcell Biochem. 2024. PMID: 38963491 Review.
-
The Interplay Between Autophagy and RNA Homeostasis: Implications for Amyotrophic Lateral Sclerosis and Frontotemporal Dementia.Front Cell Dev Biol. 2022 Apr 28;10:838402. doi: 10.3389/fcell.2022.838402. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35573690 Free PMC article. Review.
-
Role of C9orf72 hexanucleotide repeat expansions in ALS/FTD pathogenesis.Front Mol Neurosci. 2024 Jan 22;17:1322720. doi: 10.3389/fnmol.2024.1322720. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38318532 Free PMC article. Review.
-
Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.Cell. 2023 May 11;186(10):2219-2237.e29. doi: 10.1016/j.cell.2023.04.003. Cell. 2023. PMID: 37172566 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

