Heat-dependent opening of TRPV1 in the presence of capsaicin
- PMID: 34239123
- PMCID: PMC8335751
- DOI: 10.1038/s41594-021-00616-3
Heat-dependent opening of TRPV1 in the presence of capsaicin
Abstract
Transient receptor potential vanilloid member 1 (TRPV1) is a Ca2+-permeable cation channel that serves as the primary heat and capsaicin sensor in humans. Using cryo-EM, we have determined the structures of apo and capsaicin-bound full-length rat TRPV1 reconstituted into lipid nanodiscs over a range of temperatures. This has allowed us to visualize the noxious heat-induced opening of TRPV1 in the presence of capsaicin. Notably, noxious heat-dependent TRPV1 opening comprises stepwise conformational transitions. Global conformational changes across multiple subdomains of TRPV1 are followed by the rearrangement of the outer pore, leading to gate opening. Solvent-accessible surface area analyses and functional studies suggest that a subset of residues form an interaction network that is directly involved in heat sensing. Our study provides a glimpse of the molecular principles underlying noxious physical and chemical stimuli sensing by TRPV1, which can be extended to other thermal sensing ion channels.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures





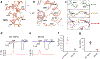
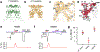
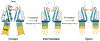
Similar articles
-
Evidence that the TRPV1 S1-S4 membrane domain contributes to thermosensing.Nat Commun. 2020 Aug 20;11(1):4169. doi: 10.1038/s41467-020-18026-2. Nat Commun. 2020. PMID: 32820172 Free PMC article.
-
Heat activation is intrinsic to the pore domain of TRPV1.Proc Natl Acad Sci U S A. 2018 Jan 9;115(2):E317-E324. doi: 10.1073/pnas.1717192115. Epub 2017 Dec 26. Proc Natl Acad Sci U S A. 2018. PMID: 29279388 Free PMC article.
-
Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid.Elife. 2017 Dec 5;6:e28626. doi: 10.7554/eLife.28626. Elife. 2017. PMID: 29206105 Free PMC article.
-
Differential effects of TRPV channel block on polymodal activation of rat cutaneous nociceptors in vitro.Exp Brain Res. 2009 Jun;196(1):31-44. doi: 10.1007/s00221-009-1808-3. Epub 2009 Apr 30. Exp Brain Res. 2009. PMID: 19404626 Review.
-
Is thermal nociception only sensed by the capsaicin receptor, TRPV1?Anat Sci Int. 2009 Sep;84(3):122-8. doi: 10.1007/s12565-009-0048-8. Epub 2009 Jun 27. Anat Sci Int. 2009. PMID: 19562439 Review.
Cited by
-
Progress in the Structural Basis of thermoTRP Channel Polymodal Gating.Int J Mol Sci. 2023 Jan 1;24(1):743. doi: 10.3390/ijms24010743. Int J Mol Sci. 2023. PMID: 36614186 Free PMC article. Review.
-
Mutagenesis studies of TRPV1 subunit interfaces informed by genomic variant analysis.Biophys J. 2023 Jan 17;122(2):322-332. doi: 10.1016/j.bpj.2022.12.012. Epub 2022 Dec 13. Biophys J. 2023. PMID: 36518076 Free PMC article.
-
TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.Sci Adv. 2024 May 3;10(18):eadn2453. doi: 10.1126/sciadv.adn2453. Epub 2024 May 1. Sci Adv. 2024. PMID: 38691614 Free PMC article.
-
Temperature Sensitive Glutamate Gating of AMPA-subtype iGluRs.bioRxiv [Preprint]. 2024 Sep 7:2024.09.05.611422. doi: 10.1101/2024.09.05.611422. bioRxiv. 2024. PMID: 39282358 Free PMC article. Preprint.
-
Structural insights into TRPV2 activation by small molecules.Nat Commun. 2022 Apr 28;13(1):2334. doi: 10.1038/s41467-022-30083-3. Nat Commun. 2022. PMID: 35484159 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

