Nucleotide Excision Repair: From Molecular Defects to Neurological Abnormalities
- PMID: 34207557
- PMCID: PMC8228863
- DOI: 10.3390/ijms22126220
Nucleotide Excision Repair: From Molecular Defects to Neurological Abnormalities
Abstract
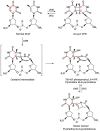
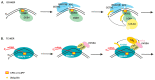
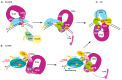
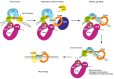
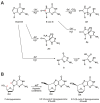
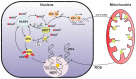
Nucleotide excision repair (NER) is the most versatile DNA repair pathway, which can remove diverse bulky DNA lesions destabilizing a DNA duplex. NER defects cause several autosomal recessive genetic disorders. Xeroderma pigmentosum (XP) is one of the NER-associated syndromes characterized by low efficiency of the removal of bulky DNA adducts generated by ultraviolet radiation. XP patients have extremely high ultraviolet-light sensitivity of sun-exposed tissues, often resulting in multiple skin and eye cancers. Some XP patients develop characteristic neurodegeneration that is believed to derive from their inability to repair neuronal DNA damaged by endogenous metabolites. A specific class of oxidatively induced DNA lesions, 8,5'-cyclopurine-2'-deoxynucleosides, is considered endogenous DNA lesions mainly responsible for neurological problems in XP. Growing evidence suggests that XP is accompanied by defective mitophagy, as in primary mitochondrial disorders. Moreover, NER pathway is absent in mitochondria, implying that the mitochondrial dysfunction is secondary to nuclear NER defects. In this review, we discuss the current understanding of the NER molecular mechanism and focuses on the NER linkage with the neurological degeneration in patients with XP. We also present recent research advances regarding NER involvement in oxidative DNA lesion repair. Finally, we highlight how mitochondrial dysfunction may be associated with XP.
Keywords: base excision repair; mitophagy; neurodegeneration; nucleotide excision repair; oxidative stress; xeroderma pigmentosum.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
The case for 8,5'-cyclopurine-2'-deoxynucleosides as endogenous DNA lesions that cause neurodegeneration in xeroderma pigmentosum.Neuroscience. 2007 Apr 14;145(4):1407-17. doi: 10.1016/j.neuroscience.2006.10.025. Epub 2006 Dec 19. Neuroscience. 2007. PMID: 17184928 Free PMC article. Review.
-
The 8,5'-cyclopurine-2'-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair.DNA Repair (Amst). 2008 Jul 1;7(7):1168-79. doi: 10.1016/j.dnarep.2008.03.016. Epub 2008 May 20. DNA Repair (Amst). 2008. PMID: 18495558 Free PMC article. Review.
-
Common pathways for ultraviolet skin carcinogenesis in the repair and replication defective groups of xeroderma pigmentosum.J Dermatol Sci. 2000 May;23(1):1-11. doi: 10.1016/s0923-1811(99)00088-2. J Dermatol Sci. 2000. PMID: 10699759 Review.
-
The cyclopurine deoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions.Free Radic Biol Med. 2017 Jun;107:90-100. doi: 10.1016/j.freeradbiomed.2016.12.028. Epub 2016 Dec 21. Free Radic Biol Med. 2017. PMID: 28011151 Review.
-
The oxidative DNA lesion 8,5'-(S)-cyclo-2'-deoxyadenosine is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells.J Biol Chem. 2000 Jul 21;275(29):22355-62. doi: 10.1074/jbc.M002259200. J Biol Chem. 2000. PMID: 10801836
Cited by
-
Does the XPA-FEN1 Interaction Concern to Nucleotide Excision Repair or Beyond?Biomolecules. 2024 Jul 9;14(7):814. doi: 10.3390/biom14070814. Biomolecules. 2024. PMID: 39062528 Free PMC article.
-
Adult-Onset Neuropsychiatric Symptoms as the Presenting Feature of Xeroderma Pigmentosum Group G: A Report of a Rare Case.Cureus. 2024 Jun 4;16(6):e61645. doi: 10.7759/cureus.61645. eCollection 2024 Jun. Cureus. 2024. PMID: 38975443 Free PMC article.
-
Skin Conditions and Movement Disorders: Hiding in Plain Sight.Mov Disord Clin Pract. 2022 Mar 24;9(5):566-583. doi: 10.1002/mdc3.13436. eCollection 2022 Jul. Mov Disord Clin Pract. 2022. PMID: 35844274 Free PMC article. Review.
-
Role of condensates in modulating DNA repair pathways and its implication for chemoresistance.J Biol Chem. 2023 Jun;299(6):104800. doi: 10.1016/j.jbc.2023.104800. Epub 2023 May 9. J Biol Chem. 2023. PMID: 37164156 Free PMC article. Review.
-
Characterization of N6-methyladenosine long non-coding RNAs in sporadic congenital cataract and age-related cataract.Int J Ophthalmol. 2024 Nov 18;17(11):1973-1986. doi: 10.18240/ijo.2024.11.02. eCollection 2024. Int J Ophthalmol. 2024. PMID: 39559306 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

