Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness
- PMID: 34183853
- PMCID: PMC7611182
- DOI: 10.1038/s41588-021-00888-x
Orphan CpG islands amplify poised enhancer regulatory activity and determine target gene responsiveness
Abstract
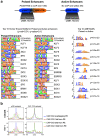
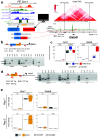
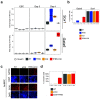
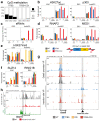
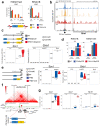
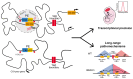
CpG islands (CGIs) represent a widespread feature of vertebrate genomes, being associated with ~70% of all gene promoters. CGIs control transcription initiation by conferring nearby promoters with unique chromatin properties. In addition, there are thousands of distal or orphan CGIs (oCGIs) whose functional relevance is barely known. Here we show that oCGIs are an essential component of poised enhancers that augment their long-range regulatory activity and control the responsiveness of their target genes. Using a knock-in strategy in mouse embryonic stem cells, we introduced poised enhancers with or without oCGIs within topologically associating domains harboring genes with different types of promoters. Analysis of the resulting cell lines revealed that oCGIs act as tethering elements that promote the physical and functional communication between poised enhancers and distally located genes, particularly those with large CGI clusters in their promoters. Therefore, by acting as genetic determinants of gene-enhancer compatibility, CGIs can contribute to gene expression control under both physiological and potentially pathological conditions.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures











Similar articles
-
Orphan CpG islands define a novel class of highly active enhancers.Epigenetics. 2017 Jun 3;12(6):449-464. doi: 10.1080/15592294.2017.1297910. Epub 2017 Apr 27. Epigenetics. 2017. PMID: 28448736 Free PMC article.
-
Frequent hypermethylation of orphan CpG islands with enhancer activity in cancer.BMC Med Genomics. 2016 Aug 12;9 Suppl 1(Suppl 1):38. doi: 10.1186/s12920-016-0198-1. BMC Med Genomics. 2016. PMID: 27534853 Free PMC article.
-
Intragenic CpG islands play important roles in bivalent chromatin assembly of developmental genes.Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):E1885-E1894. doi: 10.1073/pnas.1613300114. Epub 2017 Feb 21. Proc Natl Acad Sci U S A. 2017. PMID: 28223506 Free PMC article.
-
Sequence determinants, function, and evolution of CpG islands.Biochem Soc Trans. 2021 Jun 30;49(3):1109-1119. doi: 10.1042/BST20200695. Biochem Soc Trans. 2021. PMID: 34156435 Free PMC article. Review.
-
CpG islands and the regulation of transcription.Genes Dev. 2011 May 15;25(10):1010-22. doi: 10.1101/gad.2037511. Genes Dev. 2011. PMID: 21576262 Free PMC article. Review.
Cited by
-
Know when to fold 'em: Polycomb complexes in oncogenic 3D genome regulation.Front Cell Dev Biol. 2022 Aug 29;10:986319. doi: 10.3389/fcell.2022.986319. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36105358 Free PMC article. Review.
-
Genome organization controls transcriptional dynamics during development.Science. 2022 Feb 4;375(6580):566-570. doi: 10.1126/science.abi7178. Epub 2022 Feb 3. Science. 2022. PMID: 35113722 Free PMC article.
-
Cell fate decisions, transcription factors and signaling during early retinal development.Prog Retin Eye Res. 2022 Nov;91:101093. doi: 10.1016/j.preteyeres.2022.101093. Epub 2022 Jul 8. Prog Retin Eye Res. 2022. PMID: 35817658 Free PMC article. Review.
-
The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo.Nat Commun. 2021 Jul 16;12(1):4344. doi: 10.1038/s41467-021-24641-4. Nat Commun. 2021. PMID: 34272393 Free PMC article.
-
A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus.Dev Cell. 2021 Nov 22;56(22):3052-3065.e5. doi: 10.1016/j.devcel.2021.10.004. Epub 2021 Oct 27. Dev Cell. 2021. PMID: 34710357 Free PMC article.
References
-
- Spitz F, Furlong EEM. Transcription factors: From enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. - PubMed
-
- Kvon EZ. Using transgenic reporter assays to functionally characterize enhancers in animals. Genomics. 2015;106:185–192. - PubMed
-
- Laugsch M, et al. Modeling the Pathological Long-Range Regulatory Effects of Human Structural Variation with Patient-Specific hiPSCs. Cell Stem Cell. 2019;24:736–752.:e12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

