Upregulation of RNA cap methyltransferase RNMT drives ribosome biogenesis during T cell activation
- PMID: 34125914
- PMCID: PMC8266598
- DOI: 10.1093/nar/gkab465
Upregulation of RNA cap methyltransferase RNMT drives ribosome biogenesis during T cell activation
Abstract
The m7G cap is ubiquitous on RNAPII-transcribed RNA and has fundamental roles in eukaryotic gene expression, however its in vivo role in mammals has remained unknown. Here, we identified the m7G cap methyltransferase, RNMT, as a key mediator of T cell activation, which specifically regulates ribosome production. During T cell activation, induction of mRNA expression and ribosome biogenesis drives metabolic reprogramming, rapid proliferation and differentiation generating effector populations. We report that RNMT is induced by T cell receptor (TCR) stimulation and co-ordinates the mRNA, snoRNA and rRNA production required for ribosome biogenesis. Using transcriptomic and proteomic analyses, we demonstrate that RNMT selectively regulates the expression of terminal polypyrimidine tract (TOP) mRNAs, targets of the m7G-cap binding protein LARP1. The expression of LARP1 targets and snoRNAs involved in ribosome biogenesis is selectively compromised in Rnmt cKO CD4 T cells resulting in decreased ribosome synthesis, reduced translation rates and proliferation failure. By enhancing ribosome abundance, upregulation of RNMT co-ordinates mRNA capping and processing with increased translational capacity during T cell activation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

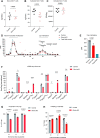
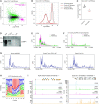
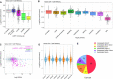

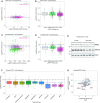
Similar articles
-
c-Myc co-ordinates mRNA cap methylation and ribosomal RNA production.Biochem J. 2017 Feb 1;474(3):377-384. doi: 10.1042/BCJ20160930. Epub 2016 Dec 1. Biochem J. 2017. PMID: 27934633 Free PMC article.
-
CDK1-Cyclin B1 Activates RNMT, Coordinating mRNA Cap Methylation with G1 Phase Transcription.Mol Cell. 2016 Mar 3;61(5):734-746. doi: 10.1016/j.molcel.2016.02.008. Mol Cell. 2016. PMID: 26942677 Free PMC article.
-
Mechanism of allosteric activation of human mRNA cap methyltransferase (RNMT) by RAM: insights from accelerated molecular dynamics simulations.Nucleic Acids Res. 2019 Sep 19;47(16):8675-8692. doi: 10.1093/nar/gkz613. Nucleic Acids Res. 2019. PMID: 31329932 Free PMC article.
-
The RNA cap methyltransferases RNMT and CMTR1 co-ordinate gene expression during neural differentiation.Biochem Soc Trans. 2023 Jun 28;51(3):1131-1141. doi: 10.1042/BST20221154. Biochem Soc Trans. 2023. PMID: 37145036 Free PMC article. Review.
-
Regulation of mRNA capping in the cell cycle.RNA Biol. 2017 Jan 2;14(1):11-14. doi: 10.1080/15476286.2016.1251540. Epub 2016 Oct 28. RNA Biol. 2017. PMID: 27791484 Free PMC article. Review.
Cited by
-
Comprehensive Analysis of m7G-Related Genes and Chronic Hepatitis B: Diagnostic Markers, Immune Microenvironment Regulation, Disease Progression.J Immunol Res. 2023 May 10;2023:9471520. doi: 10.1155/2023/9471520. eCollection 2023. J Immunol Res. 2023. PMID: 37206976 Free PMC article.
-
Biological roles of RNA m7G modification and its implications in cancer.Biol Direct. 2023 Sep 14;18(1):58. doi: 10.1186/s13062-023-00414-5. Biol Direct. 2023. PMID: 37710294 Free PMC article. Review.
-
A complementary approach for genetic diagnosis of inborn errors of immunity using proteogenomic analysis.PNAS Nexus. 2023 Mar 28;2(4):pgad104. doi: 10.1093/pnasnexus/pgad104. eCollection 2023 Apr. PNAS Nexus. 2023. PMID: 37077884 Free PMC article.
-
Identification and validation of novel genes related to immune microenvironment in polycystic ovary syndrome.Medicine (Baltimore). 2024 Oct 25;103(43):e40229. doi: 10.1097/MD.0000000000040229. Medicine (Baltimore). 2024. PMID: 39470566 Free PMC article.
-
A two-gene random forest model to diagnose osteoarthritis based on RNA-binding protein-related genes in knee cartilage tissue.Aging (Albany NY). 2023 Jan 5;15(1):193-212. doi: 10.18632/aging.204469. Epub 2023 Jan 5. Aging (Albany NY). 2023. PMID: 36641761 Free PMC article.
References
-
- Chapman N.M., Boothby M.R., Chi H.. Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 2020; 20:55–70. - PubMed
-
- Davari K., Lichti J., Gallus C., Greulich F., Uhlenhaut N.H., Heinig M., Friedel C.C., Glasmacher E.. Rapid genome-wide recruitment of RNA polymerase II drives transcription, splicing, and translation events during T cell responses. Cell Rep. 2017; 19:643–654. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

