Fluorescence Anisotropy Decays and Microscale-Volume Viscometry Reveal the Compaction of Ribosome-Bound Nascent Proteins
- PMID: 34110829
- PMCID: PMC8741338
- DOI: 10.1021/acs.jpcb.1c04473
Fluorescence Anisotropy Decays and Microscale-Volume Viscometry Reveal the Compaction of Ribosome-Bound Nascent Proteins
Abstract
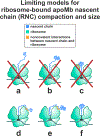
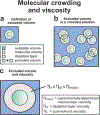
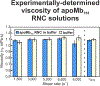
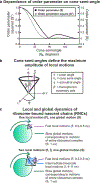
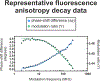
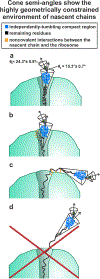
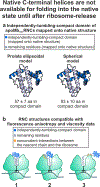
This work introduces a technology that combines fluorescence anisotropy decay with microscale-volume viscometry to investigate the compaction and dynamics of ribosome-bound nascent proteins. Protein folding in the cell, especially when nascent chains emerge from the ribosomal tunnel, is poorly understood. Previous investigations based on fluorescence anisotropy decay determined that a portion of the ribosome-bound nascent protein apomyoglobin (apoMb) forms a compact structure. This work, however, could not assess the size of the compact region. The combination of fluorescence anisotropy with microscale-volume viscometry, presented here, enables identifying the size of compact nascent-chain subdomains using a single fluorophore label. Our results demonstrate that the compact region of nascent apoMb contains 57-83 amino acids and lacks residues corresponding to the two native C-terminal helices. These amino acids are necessary for fully burying the nonpolar residues in the native structure, yet they are not available for folding before ribosome release. Therefore, apoMb requires a significant degree of post-translational folding for the generation of its native structure. In summary, the combination of fluorescence anisotropy decay and microscale-volume viscometry is a powerful approach to determine the size of independently tumbling compact regions of biomolecules. This technology is of general applicability to compact macromolecules linked to larger frameworks.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





Similar articles
-
Mapping Protein-Protein Interactions at Birth: Single-Particle Cryo-EM Analysis of a Ribosome-Nascent Globin Complex.ACS Cent Sci. 2024 Feb 1;10(2):385-401. doi: 10.1021/acscentsci.3c00777. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435509 Free PMC article.
-
Complementary Role of Co- and Post-Translational Events in De Novo Protein Biogenesis.J Phys Chem B. 2020 Jul 30;124(30):6488-6507. doi: 10.1021/acs.jpcb.0c03039. Epub 2020 Jun 9. J Phys Chem B. 2020. PMID: 32456434
-
Nanosecond dynamics of calmodulin and ribosome-bound nascent chains studied by time-resolved fluorescence anisotropy.Chembiochem. 2014 May 5;15(7):977-85. doi: 10.1002/cbic.201400014. Epub 2014 Mar 18. Chembiochem. 2014. PMID: 24644265
-
Dynamic fluorescence depolarization: a powerful tool to explore protein folding on the ribosome.Methods. 2010 Sep;52(1):57-73. doi: 10.1016/j.ymeth.2010.06.001. Epub 2010 Jun 8. Methods. 2010. PMID: 20685617 Free PMC article. Review.
-
Folding at the birth of the nascent chain: coordinating translation with co-translational folding.Curr Opin Struct Biol. 2011 Feb;21(1):25-31. doi: 10.1016/j.sbi.2010.10.008. Epub 2010 Nov 24. Curr Opin Struct Biol. 2011. PMID: 21111607 Review.
Cited by
-
Nascent chains derived from a foldable protein sequence interact with specific ribosomal surface sites near the exit tunnel.Sci Rep. 2024 May 29;14(1):12324. doi: 10.1038/s41598-024-61274-1. Sci Rep. 2024. PMID: 38811604 Free PMC article.
-
Imaging the rotational mobility of carbon dot-gold nanoparticle conjugates using frequency domain wide-field time-resolved fluorescence anisotropy.J Biomed Opt. 2023 May;28(5):056001. doi: 10.1117/1.JBO.28.5.056001. Epub 2023 May 23. J Biomed Opt. 2023. PMID: 37229274 Free PMC article.
-
Critical Beginnings: Selective Tuning of Solubility and Structural Accuracy of Newly Synthesized Proteins by the Hsp70 Chaperone System.J Phys Chem B. 2023 May 11;127(18):3990-4014. doi: 10.1021/acs.jpcb.2c08485. Epub 2023 May 2. J Phys Chem B. 2023. PMID: 37130318 Free PMC article.
-
Mapping Protein-Protein Interactions at Birth: Single-Particle Cryo-EM Analysis of a Ribosome-Nascent Globin Complex.ACS Cent Sci. 2024 Feb 1;10(2):385-401. doi: 10.1021/acscentsci.3c00777. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435509 Free PMC article.
-
Protein folding in vitro and in the cell: From a solitary journey to a team effort.Biophys Chem. 2022 Aug;287:106821. doi: 10.1016/j.bpc.2022.106821. Epub 2022 Apr 29. Biophys Chem. 2022. PMID: 35667131 Free PMC article. Review.
References
-
- Dill KA; MacCallum JL The protein-folding problem, 50 years on. Science 2012, 338, 1042–1046. - PubMed
-
- Udgaonkar JB Polypeptide chain collapse and protein folding. Arch. Biochem. Biophys. 2013, 531, 24–33. - PubMed
-
- Balchin D; Hayer-Hartl M; Hartl FU In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

